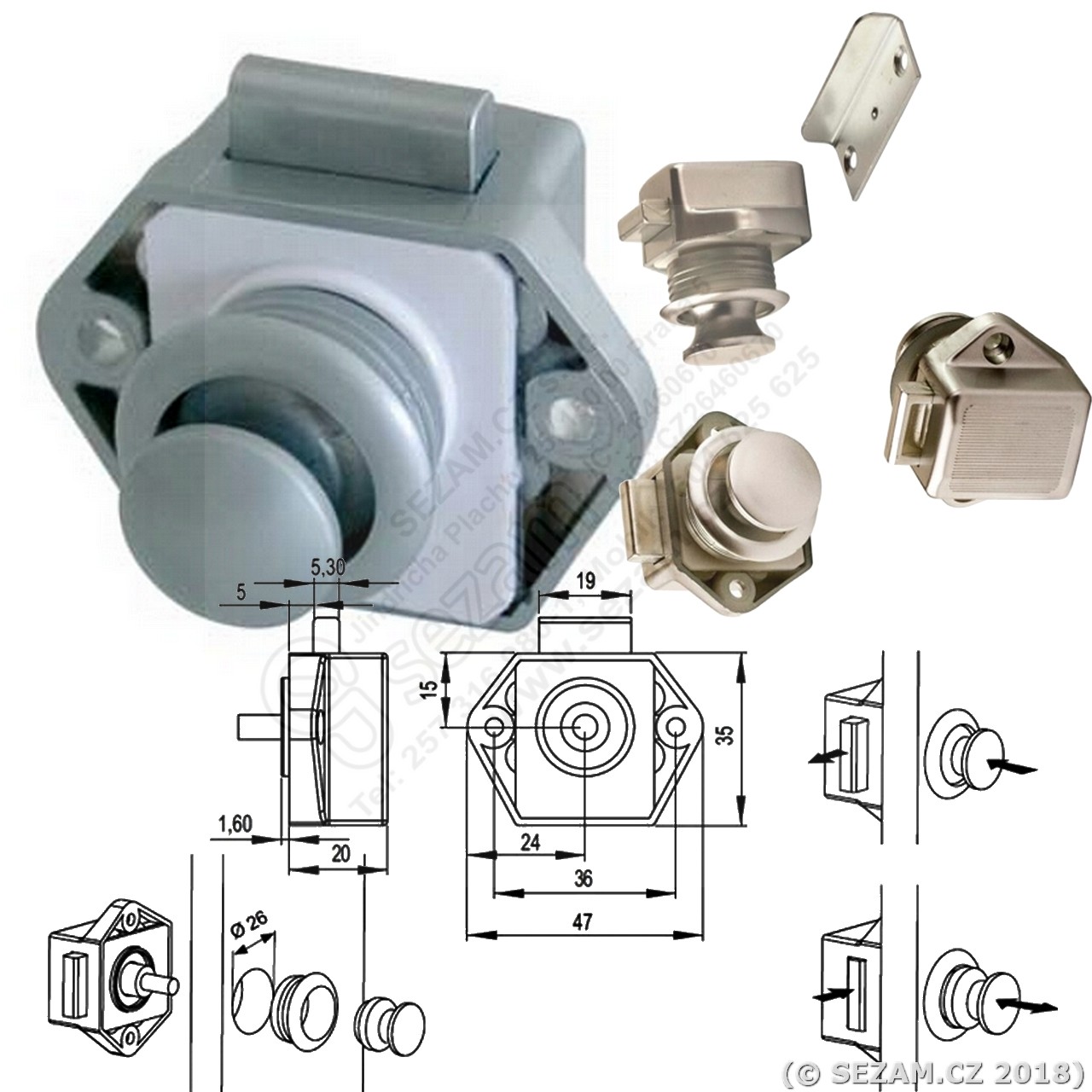
Cílem Rv Karavan Vstupu Zámku Dveří Karavan Západky Dveří Rukojeť Klíče Rv Zamykání Náhradní Pro Rv/karavan/přívěs - Velkoobchod \ www.senatordoubrava.cz

Koupit Rv Wc Koupelna Zámku Dveří Karavan, Auto, Karavan, Loď Zvládnout Knoflík Kabinet šuplíku Zámek Push Button Dveřní Zámky Pro Nábytek \ Automobily, Díly A Příslušenství ~ www.casuntour.cz