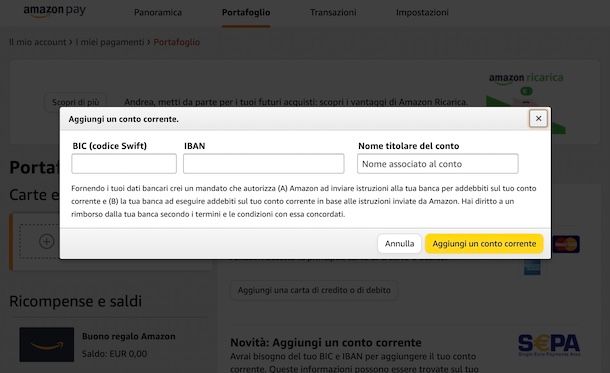
Foglio Informativo e Documento di Sintesi delle Carte Amazon Business American Express® e Carte Amazon Business Prime American

Lonnalee Blocco di Protezione RFID, Set da 10 | Schermatura Carta di Credito | Proteggi Carte di Credito Contactless(Colorato) : Amazon.it: Moda


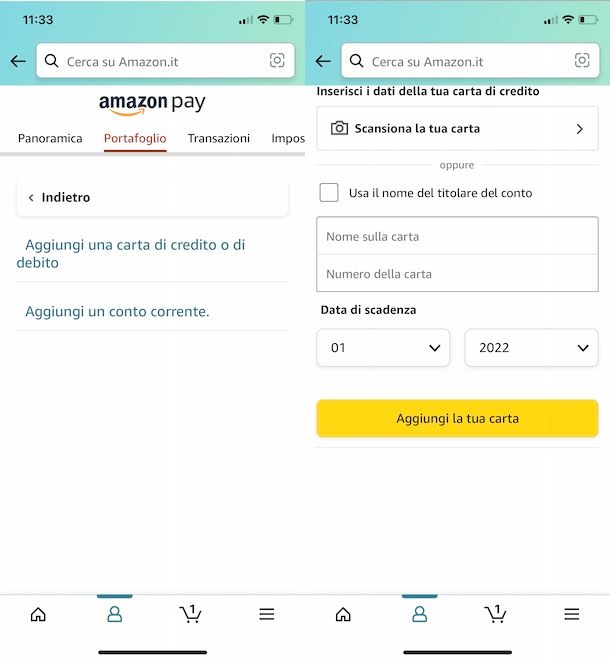




![Revisione della carta di credito Amazon nel 2023 [testato da esperti] Revisione della carta di credito Amazon nel 2023 [testato da esperti]](https://flyingcdn-752ef22c.b-cdn.net/wp-content/uploads/2023/02/What-Is-The-Amazon-Credit-Card-Credit-Limit.jpg?width=800)









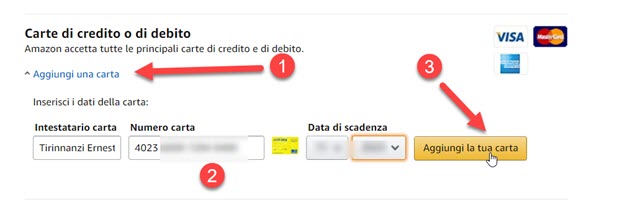
![Revisione della carta di credito Amazon nel 2023 [testato da esperti] Revisione della carta di credito Amazon nel 2023 [testato da esperti]](https://flyingcdn-752ef22c.b-cdn.net/wp-content/uploads/2023/02/What-Is-An-Amazon-Credit-Card.jpg)