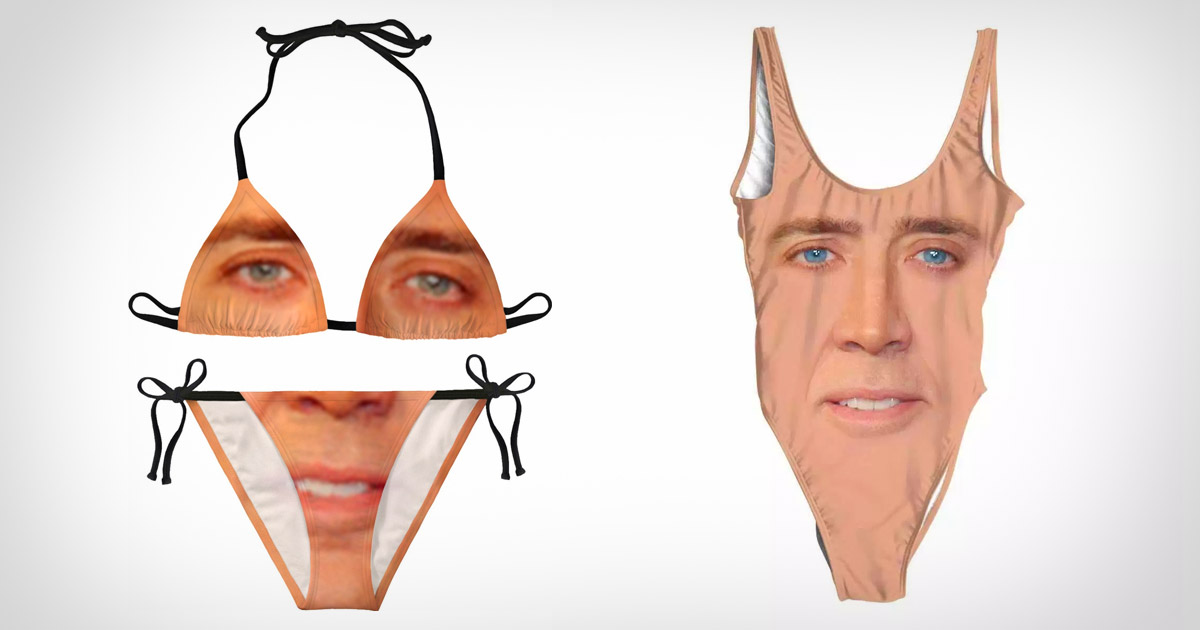
jesse@metasocial.com - I don't post here anymore on Twitter: "Nicholas Cage face swimsuit. https://t.co/3zlHfUvOWu I wonder how many more of these Face/Off tie-ins there are on Aliexpress https://t.co/kKfeSgrmkN" / Twitter

Kate Upton Sports Illustrated swimsuit cover revealed, Nicolas Cage wants to be like Led Zeppelin, and more - pennlive.com

https://www.rageon.com/products/nicolas-cage-bikini-1 / Nicolas Cage :: bikini / смешные картинки и другие приколы: комиксы, гиф анимация, видео, лучший интеллектуальный юмор.

Nicholas Cage Swimsuit | Nicolas Cage Swimsuit | Nicolas Cage Swimwear | Suit Nicholas 1 - One-piece Suits - Aliexpress

Shocked' Trump face, Nicolas Cage, luchador and many more WEIRD one-piece swimsuits | Dangerous Minds

Nicolas Cage & Alice Kim: Magical Mates: Photo 2464416 | Alice Kim, Nicolas Cage Photos | Just Jared: Entertainment News

GloryThePen on Twitter: "@lazyspacebear I see your minion bikini and raise you a thanos bikini https://t.co/yIbQ0s8h0v" / Twitter

Nicolas Cage Bikini Swimsuit Funny Meme Print High Cut Swimwear Trendy Bikini Set Push Up Graphic Feminine Bikinis Beach Wear - Bikinis Set - AliExpress