Jazda bez uprawnień, bez odpowiedniej kategorii 2023. Jaki mandat za brak motocyklowego prawa jazdy AM, A1, A2, A | Motovoyager

Wniosek do sądu za jazdę bez uprawnień i mandat za udostępnienie pojazdu osobie bez prawa jazdy - Informacje - Komenda Powiatowa Policji w Rawie Mazowieckiej

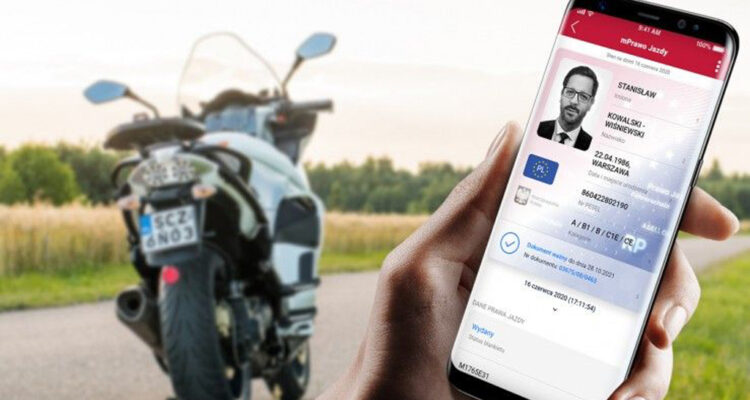
Jazda bez prawa jazdy od 5 grudnia. Nie trzeba wozić ze sobą dokumentu - Jaworzno - Portal Społecznościowy - jaw.pl