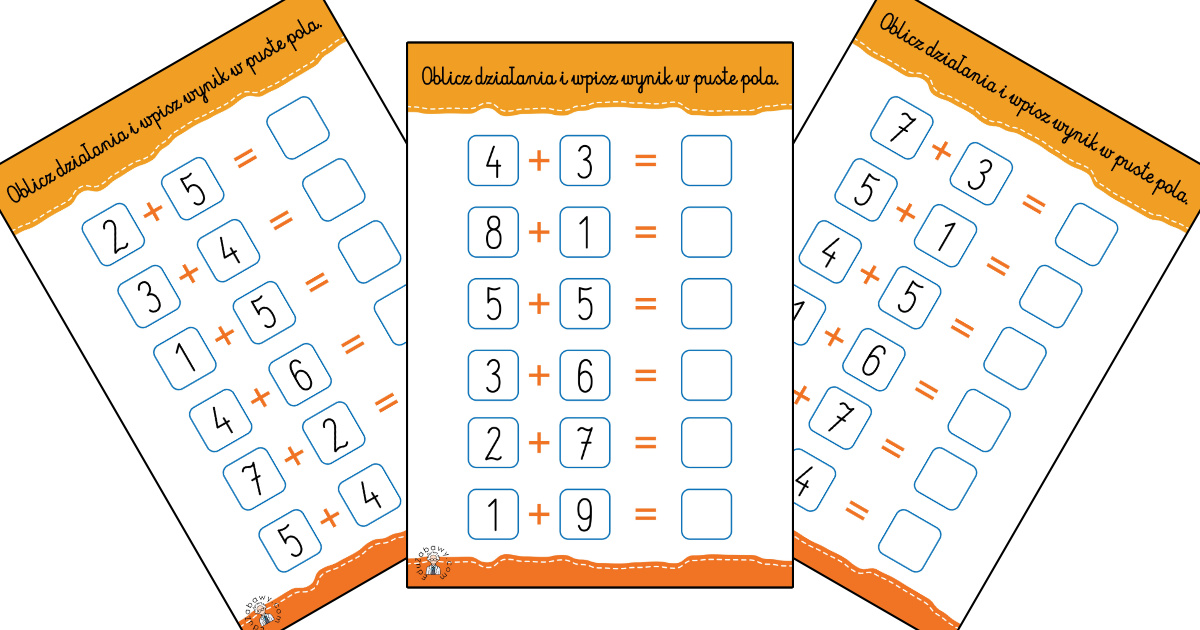
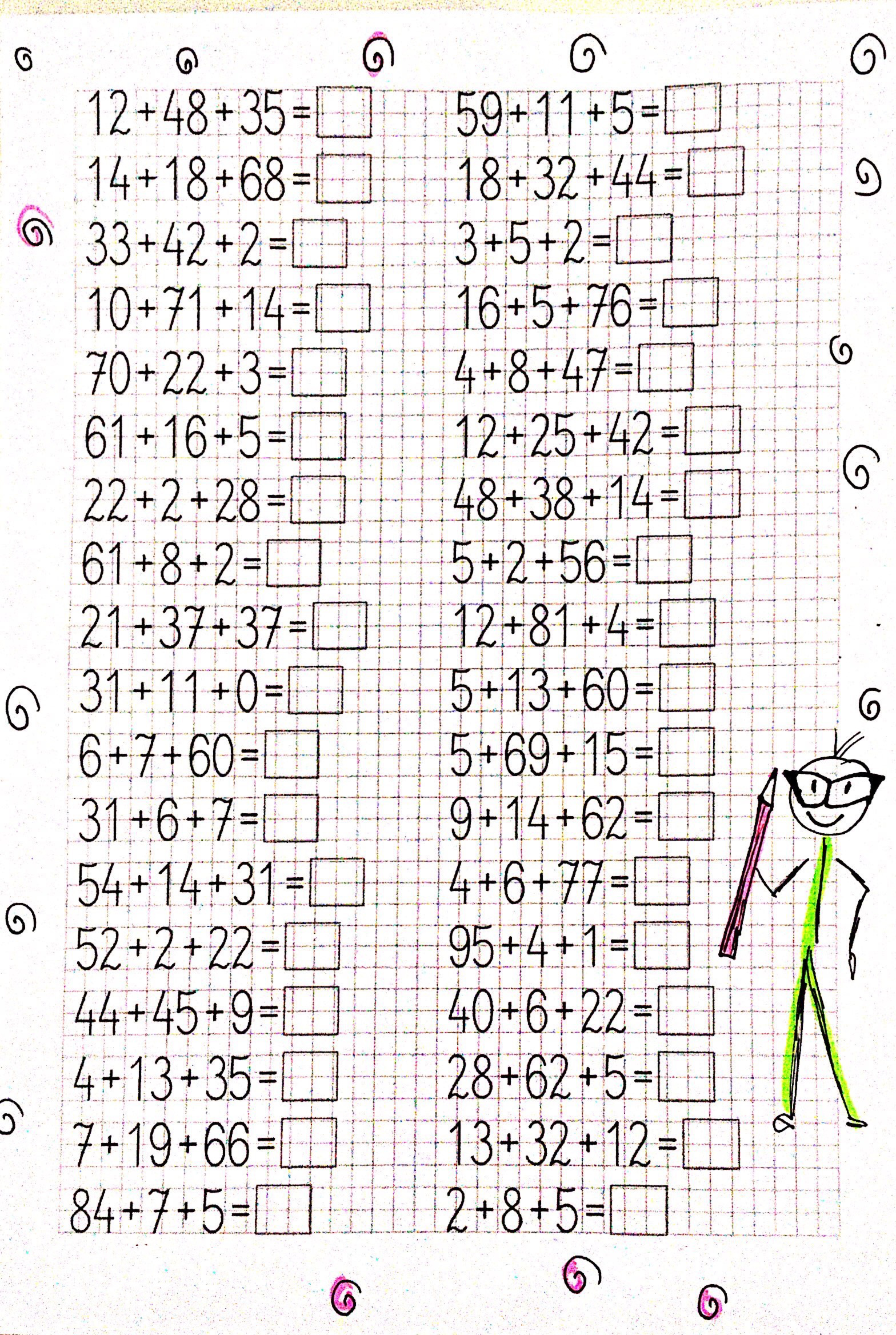
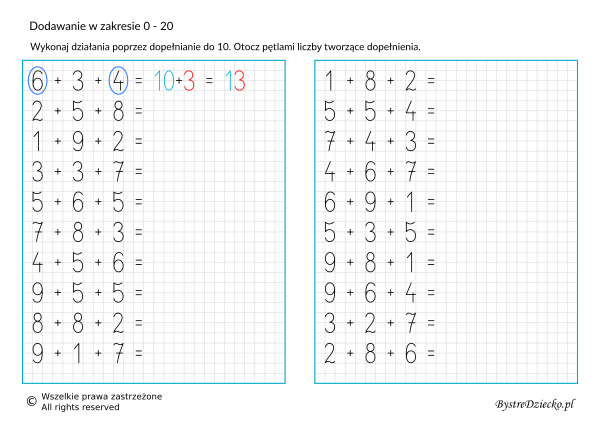
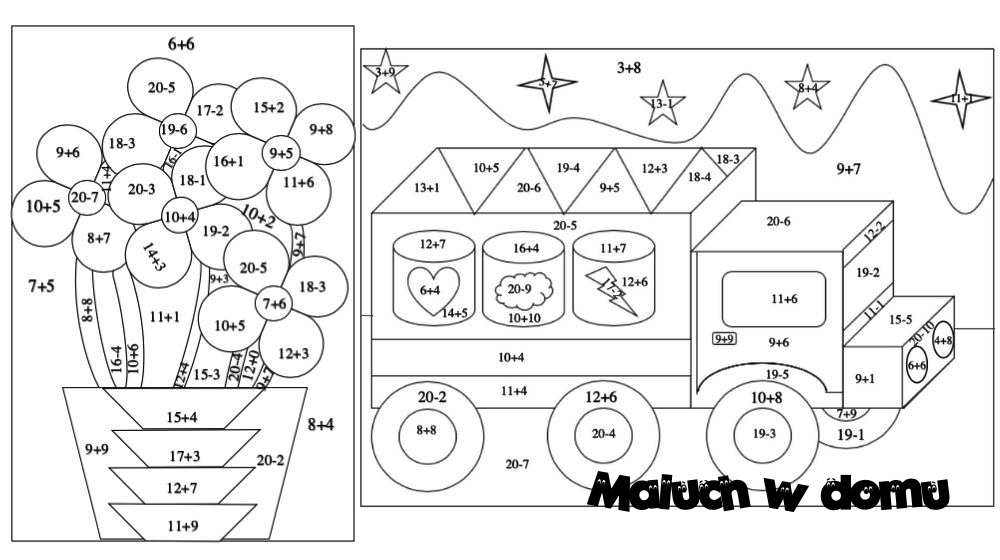
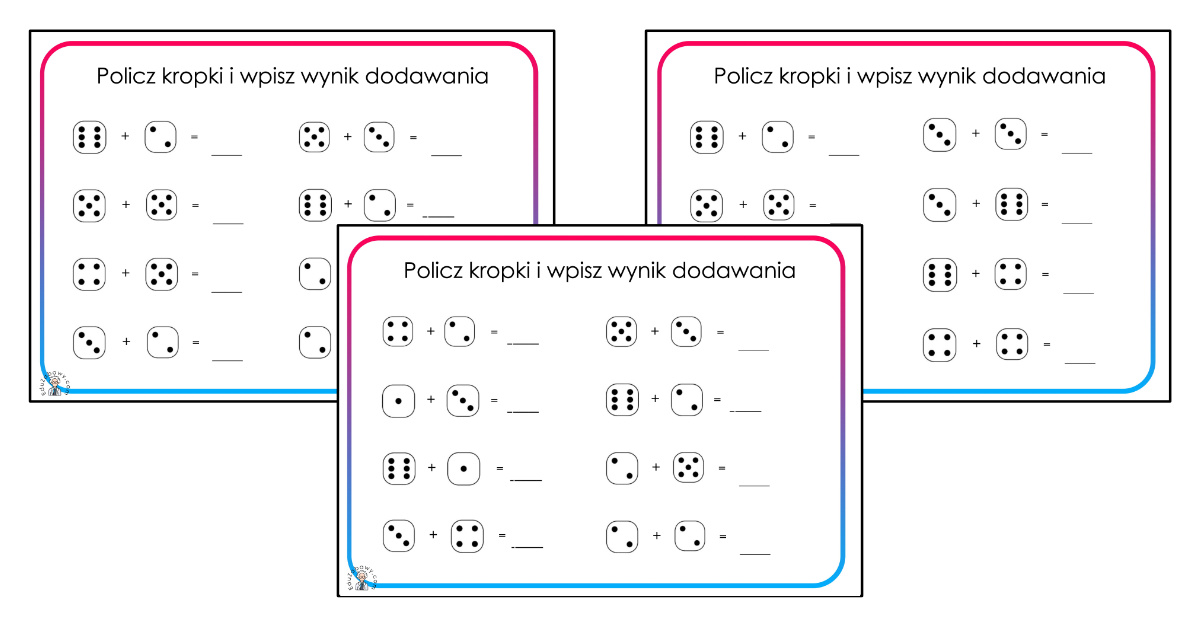
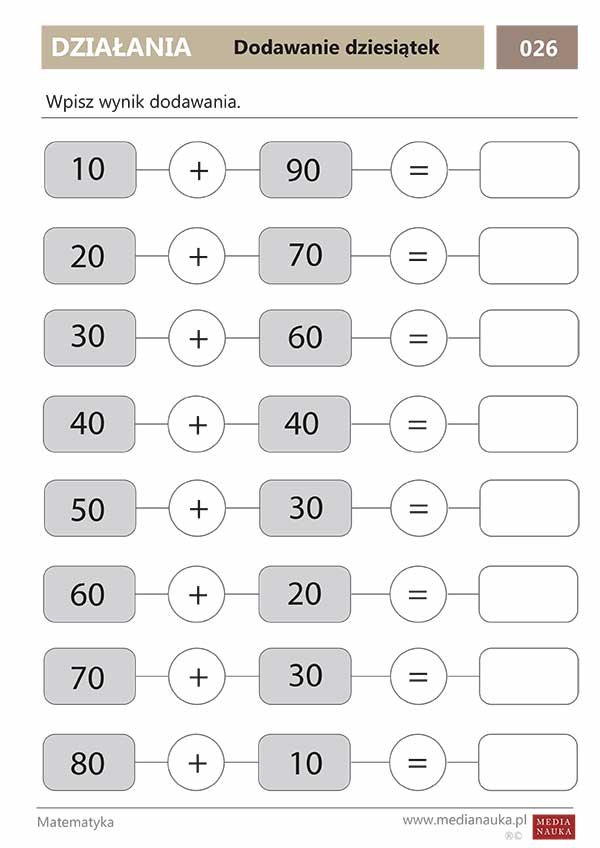
Dodawanie w zakresie 10 – karty pracy dla dzieci - Bystre Dziecko | Math for kids, Kids math worksheets, Math

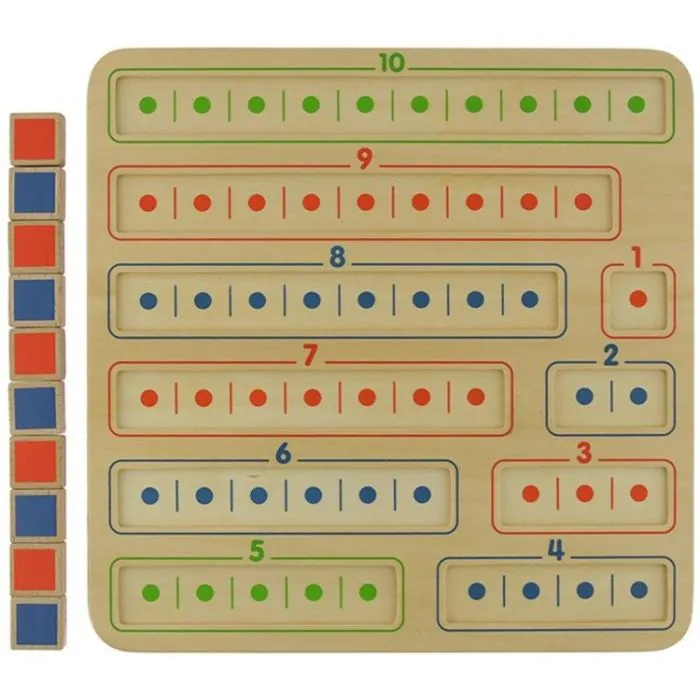
Nauka Dodawania I Odejmowania Tabliczka Edukacyjna Masterkidz - Zabawki rozwojowe - ZABAWKI - Ceny i opinie - Maludas.pl
![Matematyka, Zadania] Dodawanie w zakresie 10 | Math worksheets, Kindergarten math worksheets, Alphabet kindergarten Matematyka, Zadania] Dodawanie w zakresie 10 | Math worksheets, Kindergarten math worksheets, Alphabet kindergarten](https://i.pinimg.com/736x/86/44/b1/8644b1dd0fccdd81b23c1447e39683c7.jpg)
Matematyka, Zadania] Dodawanie w zakresie 10 | Math worksheets, Kindergarten math worksheets, Alphabet kindergarten