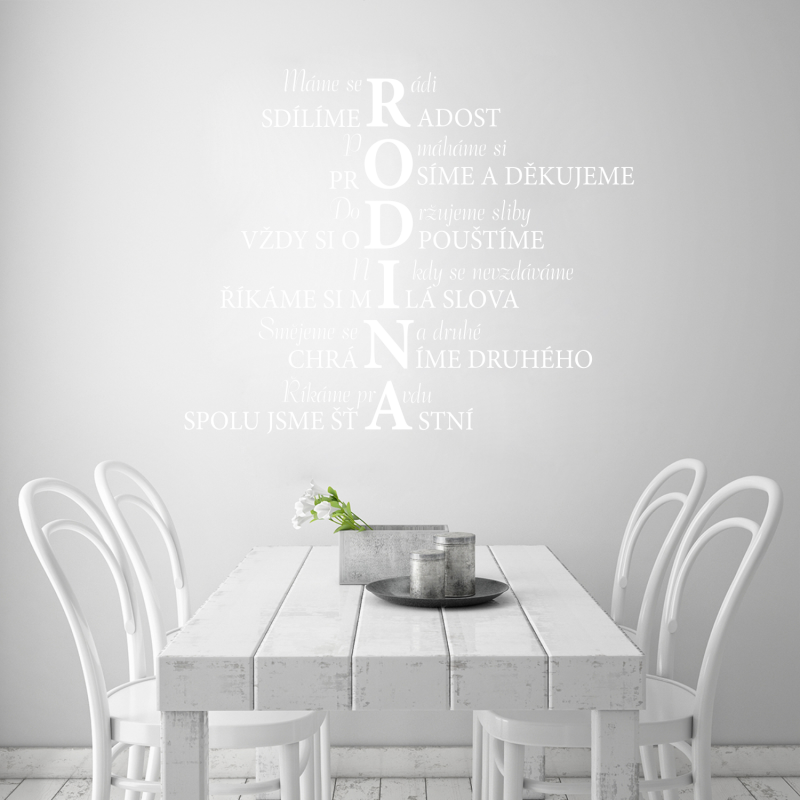
SaMolepka na zeď - Mít místo, kam se můžu vrátit je domov. Mít někoho, kdo Tě doma miluje, je rodina. Mít oboje je dar. | Gravon.cz

Samolepky na zeď, dekorace na zeď, nálepky na zeď, samolepicí dekorace na zeď, moderní samolepky na zeď, citáty, nápisy, | AZsamolepky.cz
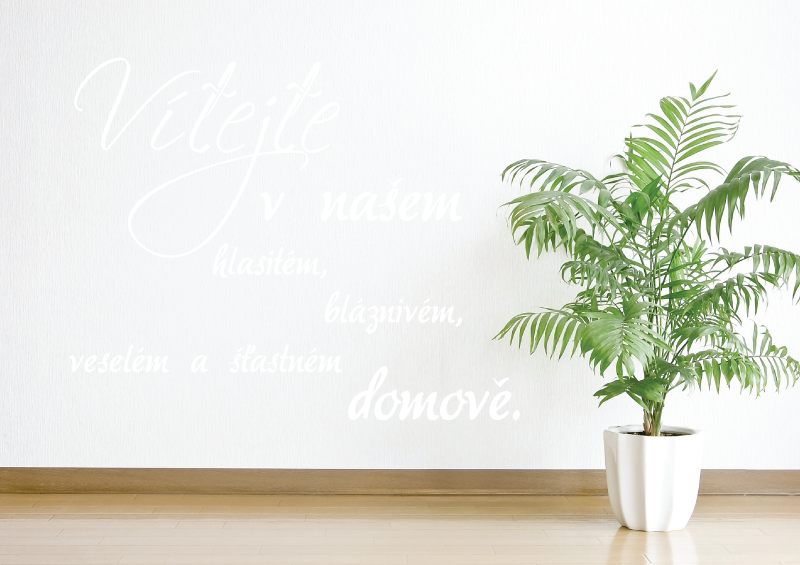
Samolepky na zeď, dekorace na zeď, nálepky na zeď, samolepicí dekorace na zeď, moderní samolepky na zeď, texty, nápisy, | AZsamolepky.cz

Samolepky na zeď, dekorace na zeď, nálepky na zeď, samolepicí dekorace na zeď, moderní samolepky na zeď, nápis, citát, d | AZsamolepky.cz
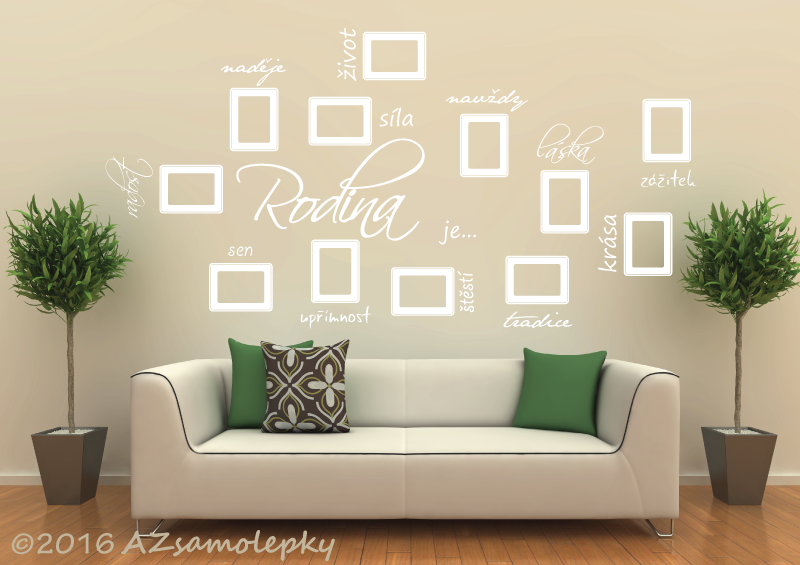
Samolepky na zeď, dekorace na zeď, nálepky na zeď, samolepicí dekorace na zeď, samolepicí fotorámečky na zeď, nápis, rod | AZsamolepky.cz

Samolepky na zeď, dekorace na zeď, nálepky na zeď, samolepicí dekorace na zeď, nápis, citát, text, vejdi | AZsamolepky.cz