
KASMOM 3 мм, м Тел за Бижута направи си Алуминиева Ремесленная Тел Сребро Злато Бижута Www.savingprimates.org

Алуминиева тел за изработката на бижута колие направи си сам гривна 0,8 мм 1 мм, 1,5 мм 2 мм 3 мм, меки мъниста бонсай цветен панела за бродерия тел купи онлайн <

Hotop 5 Ролки Тел Гъвкава Анодизиран Ремесленная за Изработката Бижута Мека Метална Жица за направи Savingprimates.org

10 м/ролка 5x1 mm 12 цвята плоска алуминиева мека метална тел за бижута направи си сам crafts кабел | Изработка на мъниста и Бижута > Untreasure.co.uk

5 ролки 3 мм 5 мм плоска алуминиева тел, ръчно изработени занаяти метална тел за бижута направи си сам огърлица, гривна, обеци и аксесоари Разпродажба < Изработка на мъниста и бижута \ www.dividendmax.eu

0,2-1 мм beading златен посеребренный меден кабел Низ конец Метална тел за колиета Гривни Аксесоари за бижута направи си САМ купи / Изработка на мъниста и бижута \ www.dealers-skoda.co.uk

0,2-1 мм цвят медна тел за гривни, колиета, бижута, бисероплетения, метална, сребро/злато/розово злато, търговия на едро купи онлайн | Изработка на мъниста и бижута \ www.haaratelier-manuela.app
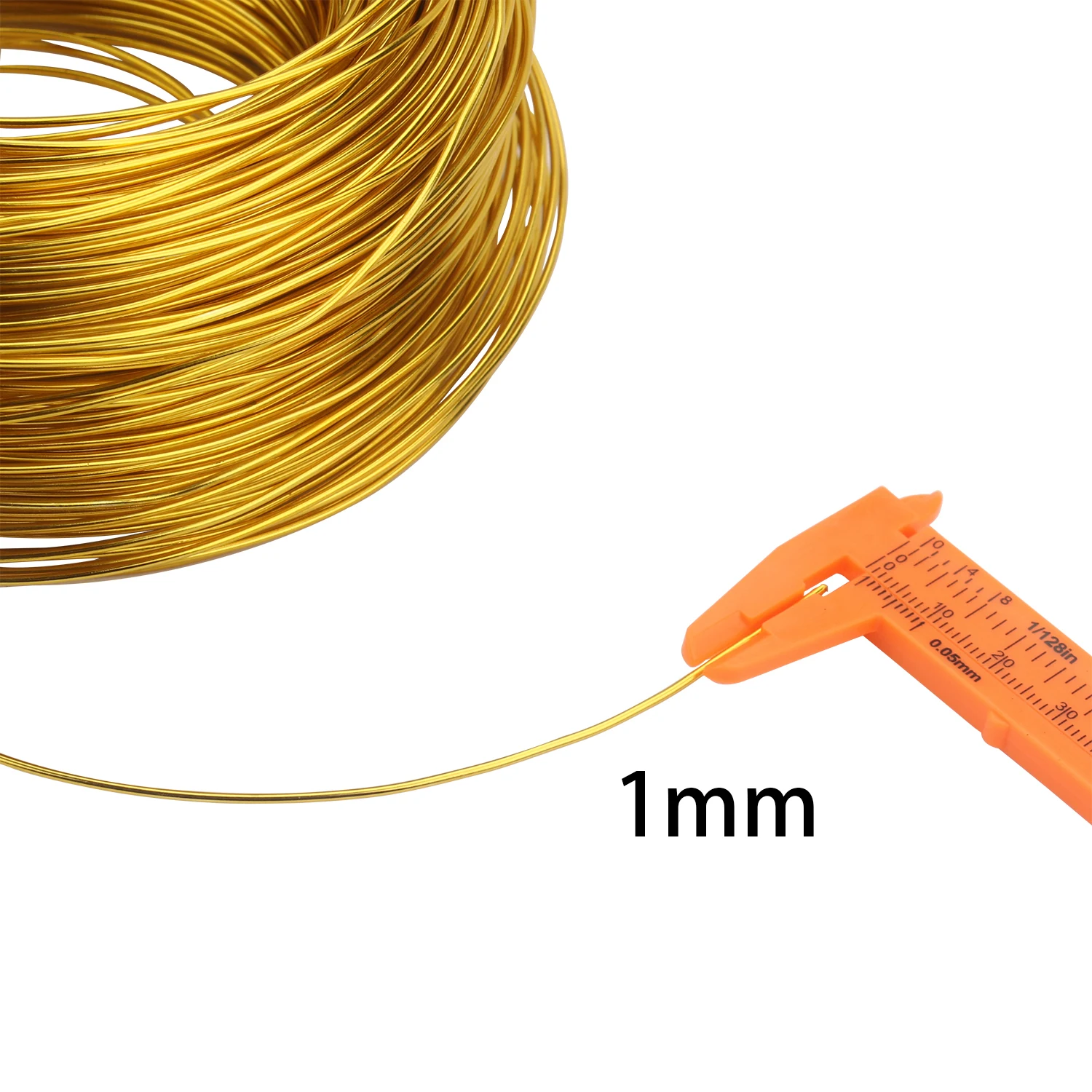
Бижута, аксесоари Позлатен Алуминиева Тел 1 мм/1.2 мм/1.5 мм/2 мм/2.5 мм/3 мм Алуминиева Метална Тел за бижута направи си САМ Разпродажба \ Изработка на мъниста и бижута ~ www.iplogger.shop

3 мм, 4 мм, 25/12 м алуминиева тел бижута изводи за бижута направи си сам алуминий занаят тел сребро злато бижута лека метална тел купи онлайн > Изработка На Мъниста И Бижута \ Sales-Original.cam

Бижута, аксесоари Позлатен Алуминиева Тел 1 мм/1.2 мм/1.5 мм/2 мм/2.5 мм/3 мм Алуминиева Метална Тел за бижута направи си САМ Разпродажба \ Изработка на мъниста и бижута ~ www.iplogger.shop
















