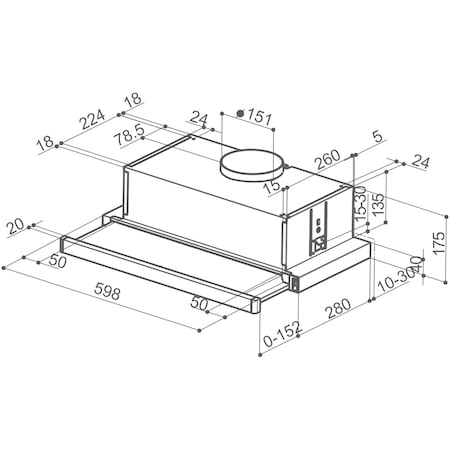
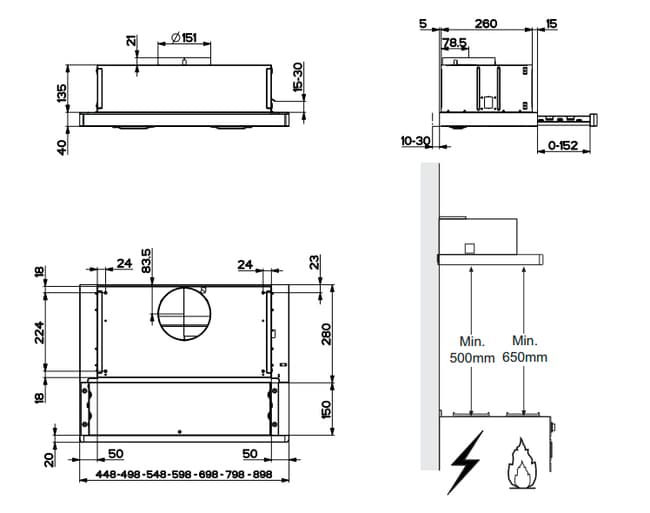
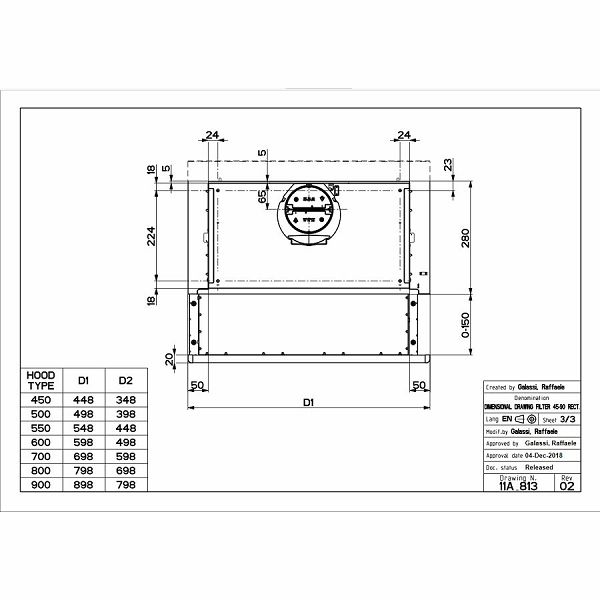
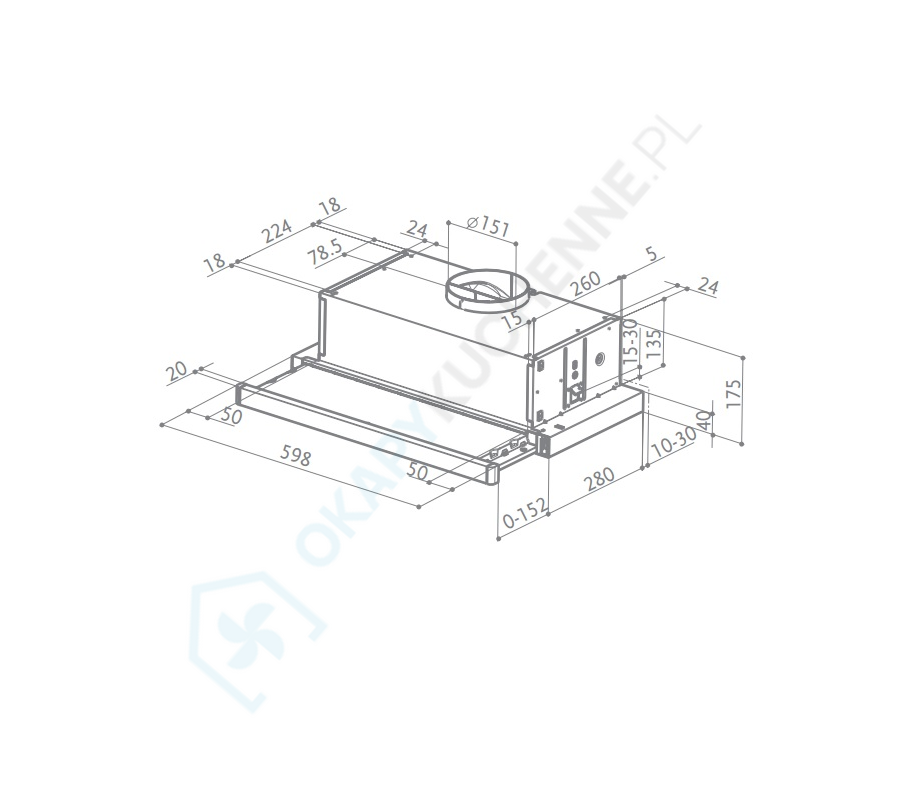
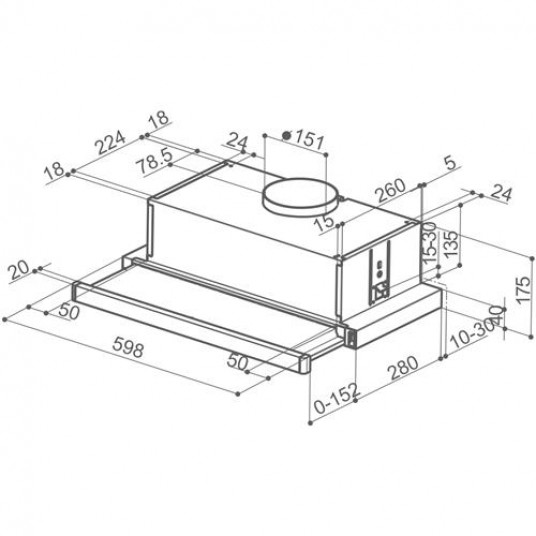
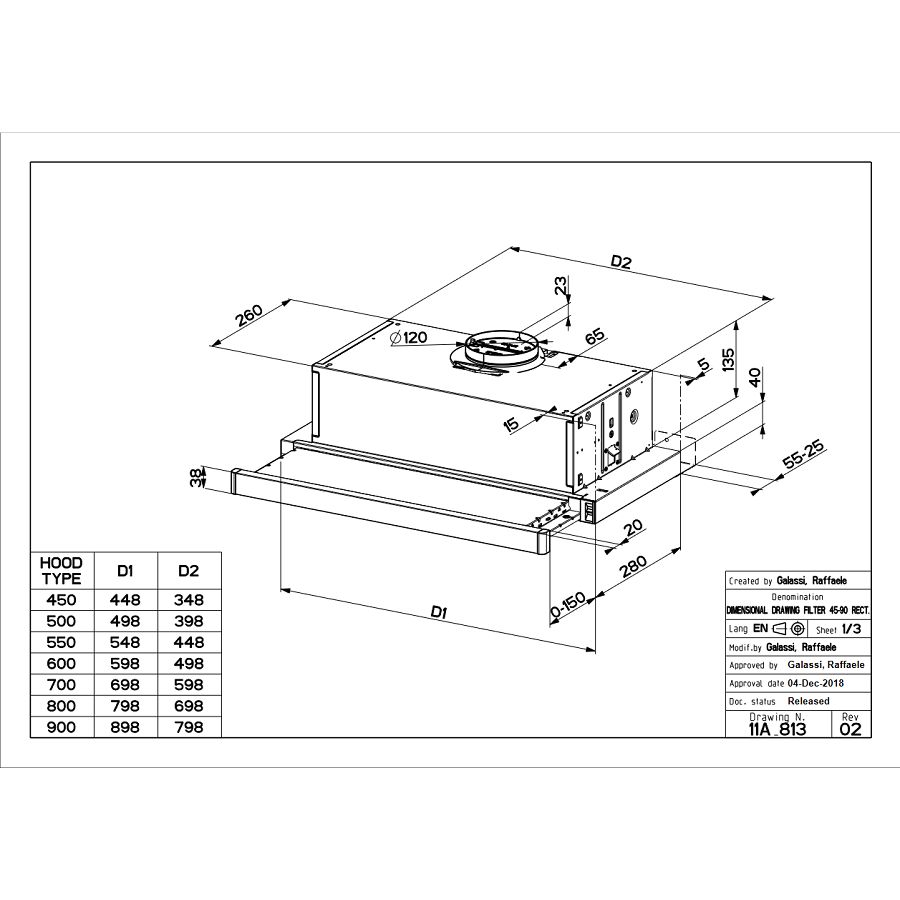
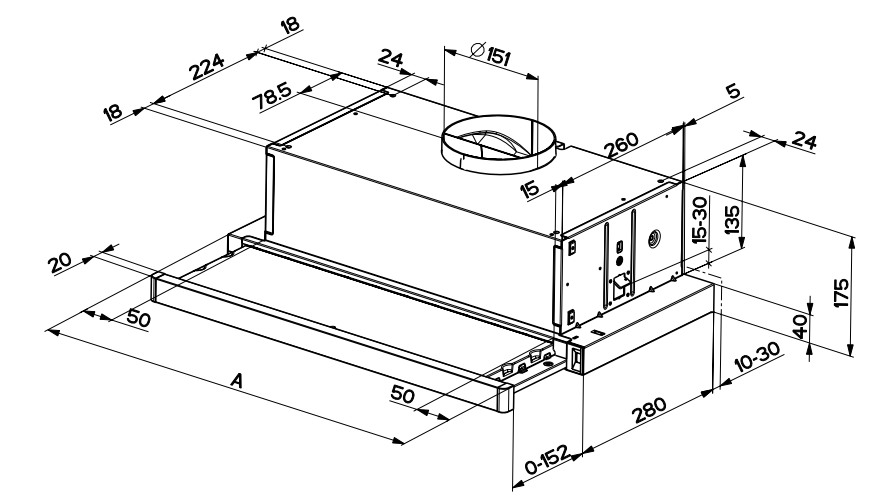
Вытяжка «Faber» Flexa glass, M6 W A60, 110.0294.082 купить с доставкой, цены в интернет-магазине Едоставка

Цена на Faber FLEXA GLASS M6 W A60 - 10990 руб в Москве, купить с бесплатной доставкой вытяжку Faber FLEXA GLASS M6 W A60 прочитав отзывы, описания и инструкции на Hausdorf
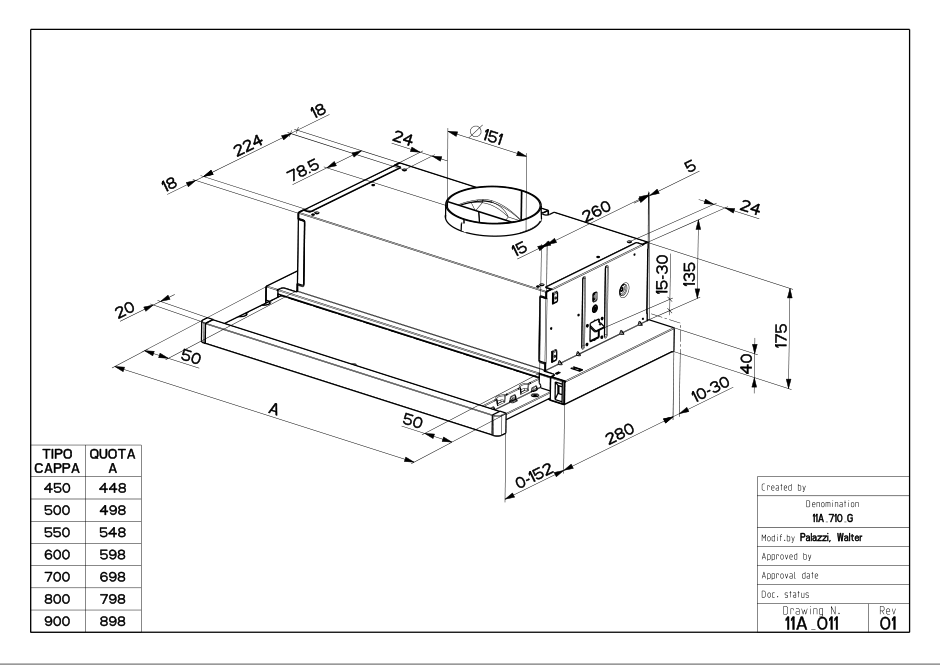
Встраиваемая вытяжка Faber FLEXA GLASS M6 W A60 в фирменном магазине Фабер. Официальная гарантия, доставка по России от Faber
Цена на Faber FLEXA GLASS M6 W A60 - 10990 руб в Москве, купить с бесплатной доставкой вытяжку Faber FLEXA GLASS M6 W A60 прочитав отзывы, описания и инструкции на Hausdorf