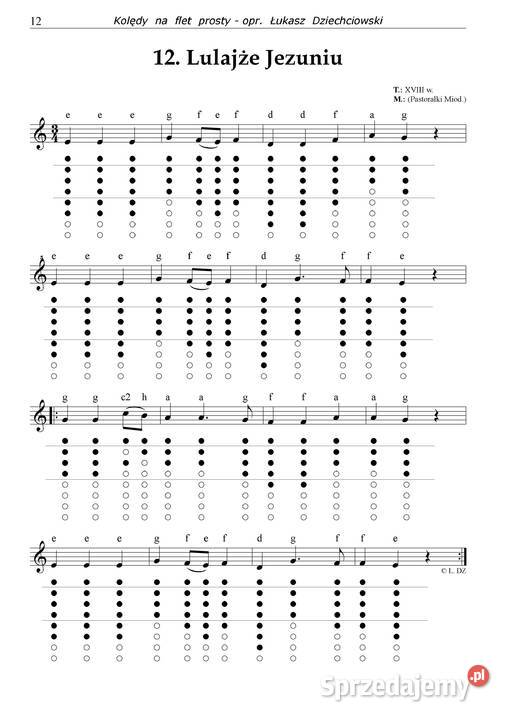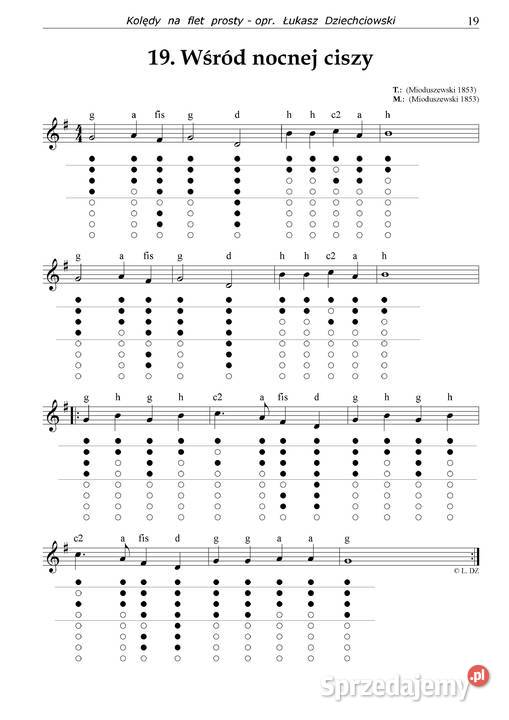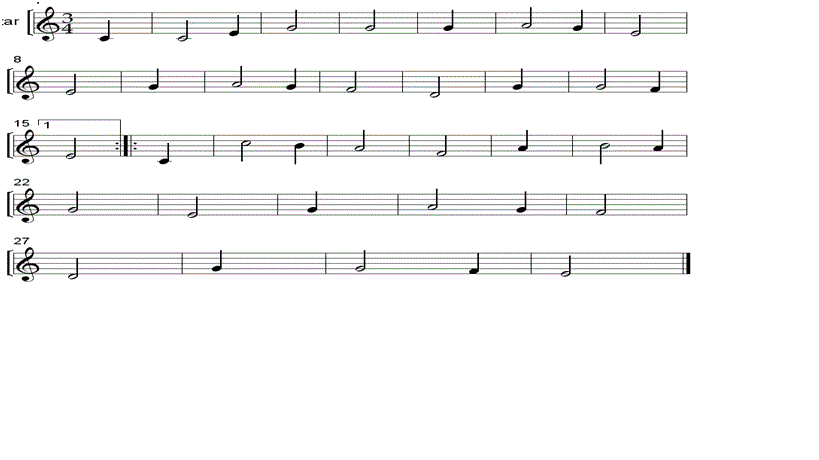
74: Strona główna - Nuty na flet prosty - utwory obowiązkowe | Szkoła Podstawowa im.Powstańców Wielkopolskich

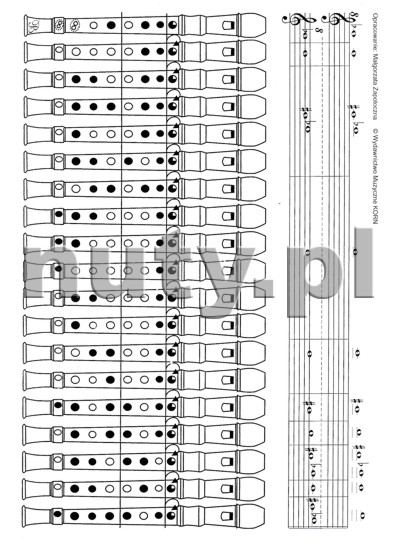
Lőrincz László, Paragi Jenő - Recorder ABC for C descant recorder - na flet prosty sopranowy z akompaniamentem fortepianu MUZO

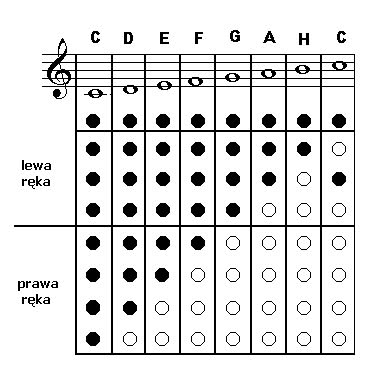
Bardzo proszę chwyty na flet prosty Arie Skobuły mam zaliczenie a niewiem jak to zagrac .Proszę pomóżcie - Brainly.pl
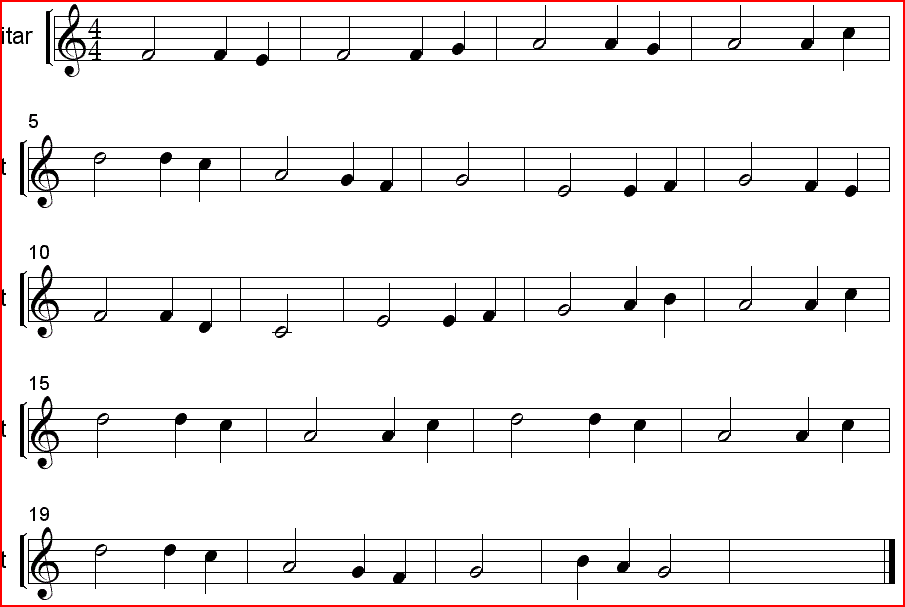
74: Strona główna - Nuty na flet prosty - utwory obowiązkowe | Szkoła Podstawowa im.Powstańców Wielkopolskich

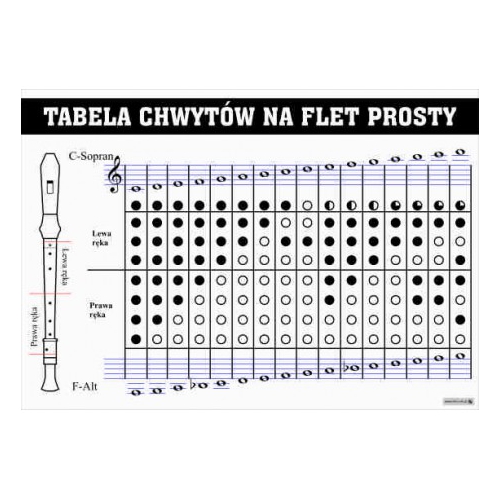
Dzień dobry potrzebuje żeby ktoś mi napisał nuty do fletu renesansowego i przetłumaczyć na chwyty dziękuje z - Brainly.pl