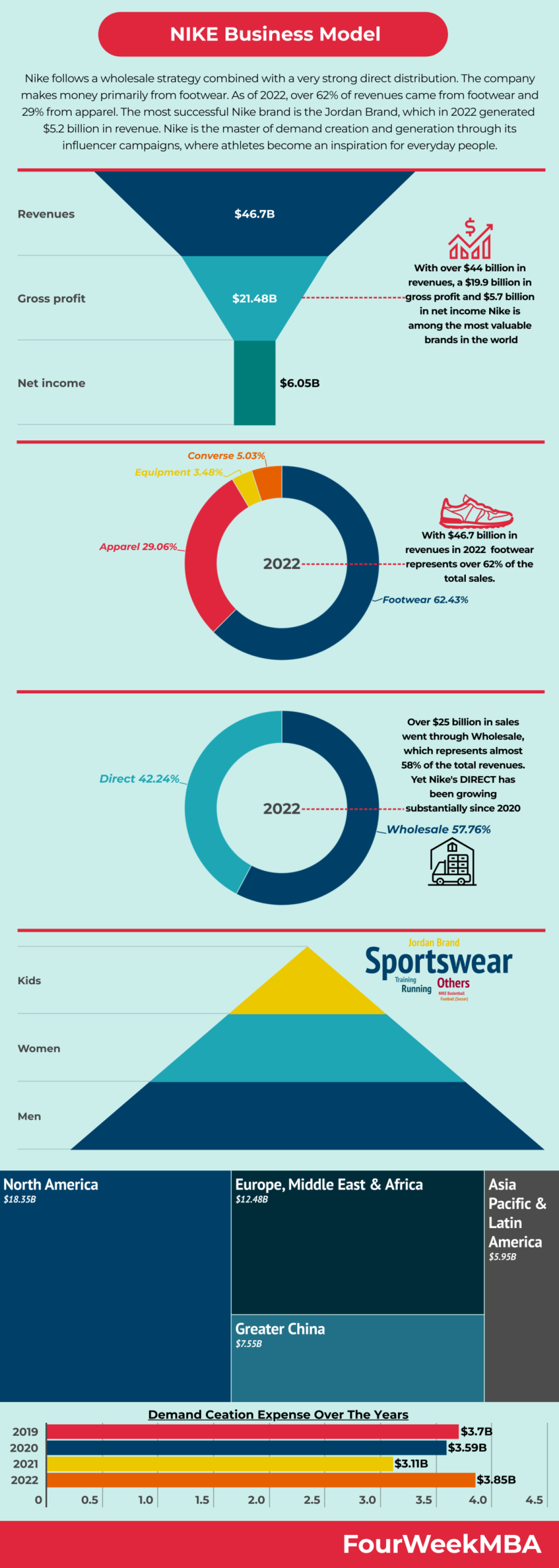
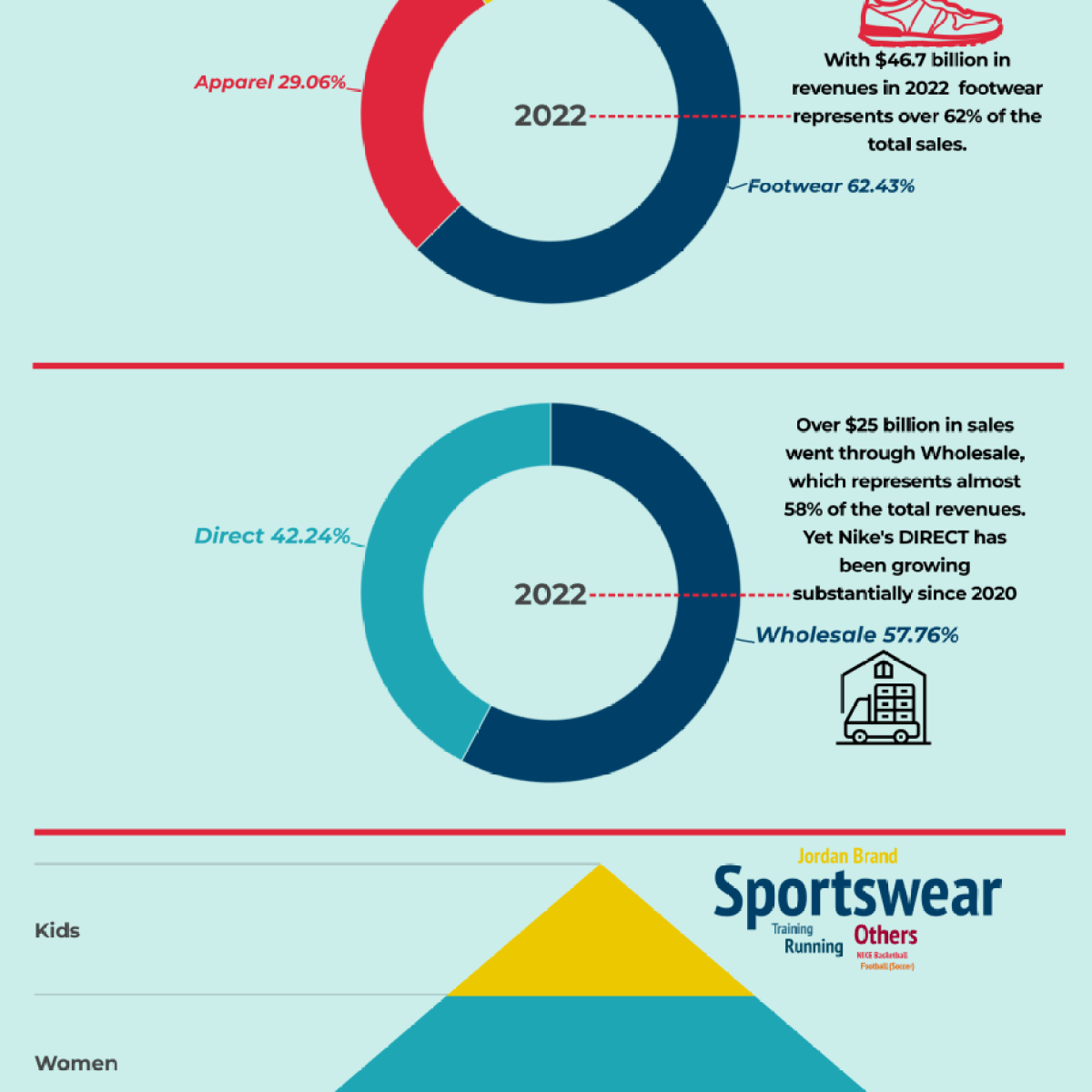
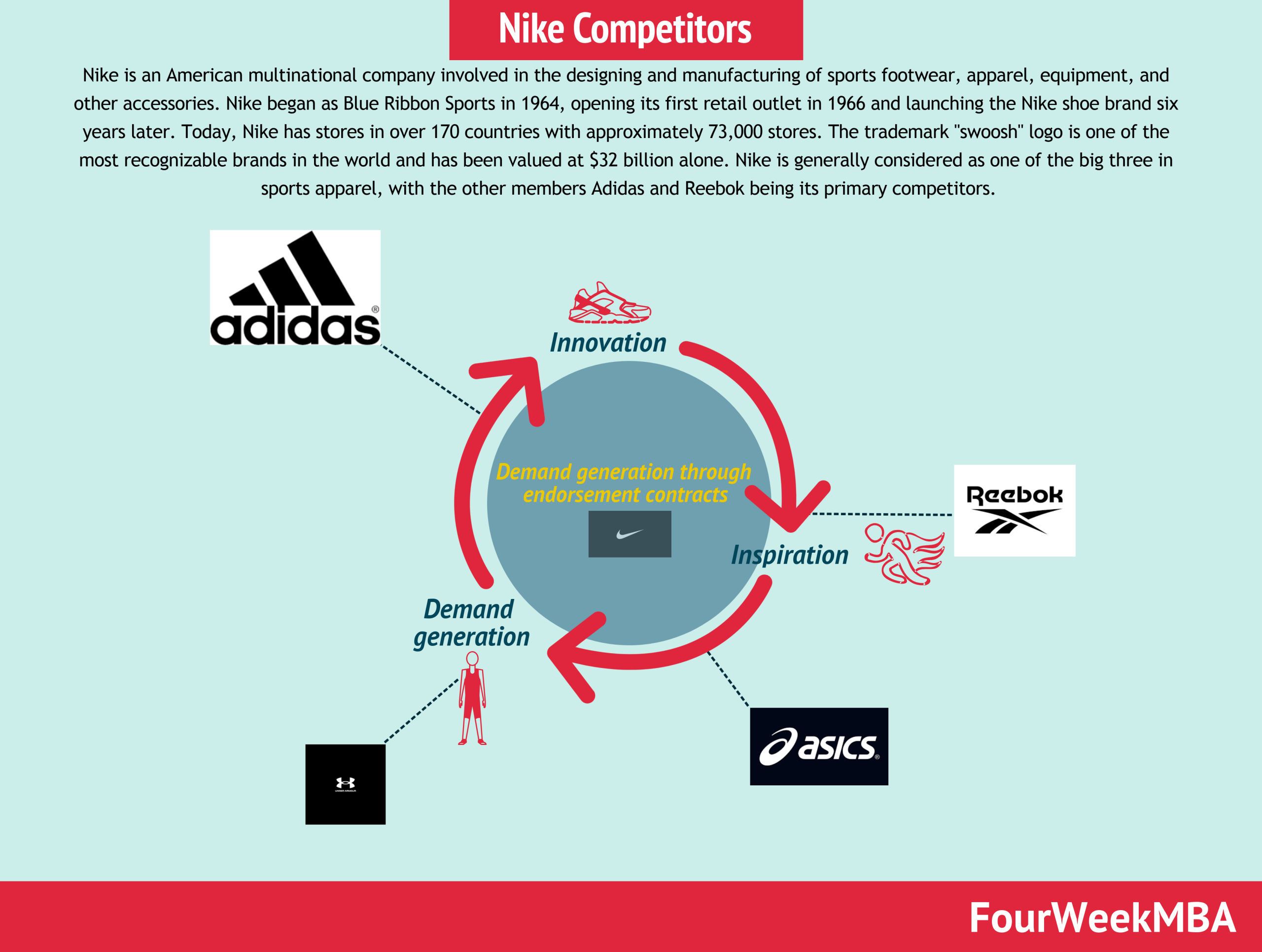
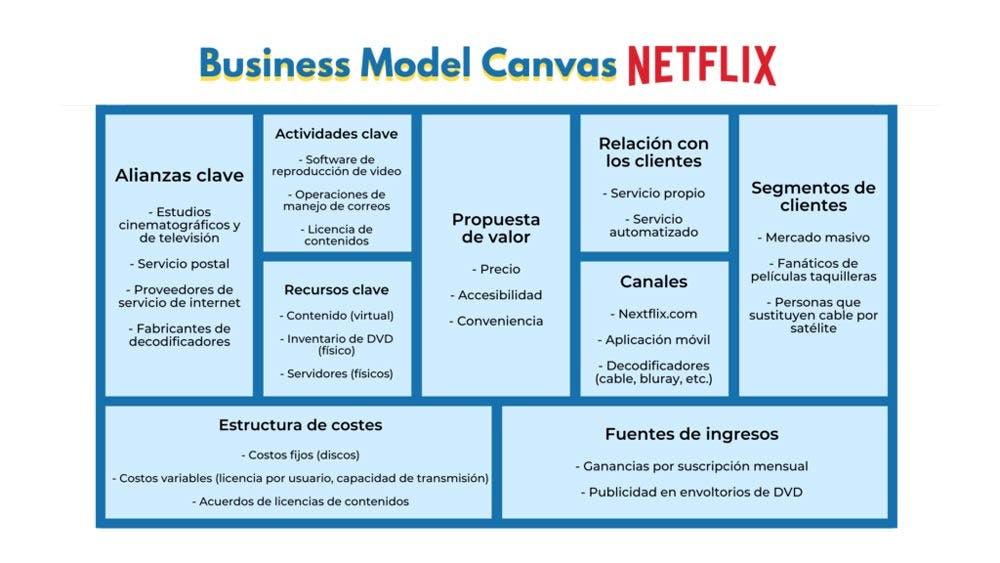
Modelo Canvas Nike.docx - Modelo de Negocio Canvas de Nike Socios claves Converse Pony Hurley International Actividades claves Ofrecer ropa y calzado | Course Hero

Nike confirmó que se va del país y le deja su negocio a un licenciatario internacional - FM POWER RIO TERCERO

Modelo Canvas Nike.docx - Modelo de Negocio Canvas de Nike Socios claves Converse Pony Hurley International Actividades claves Ofrecer ropa y calzado | Course Hero