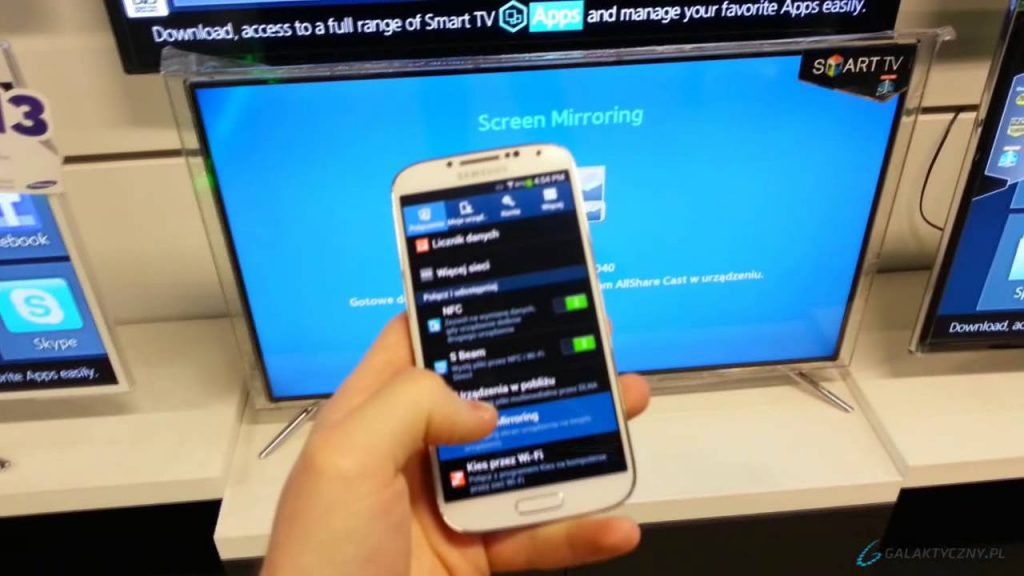
How to Start Remote Control Computer and Screen Mirroring in SAMSUNG A310F Galaxy A3 (2016)? - HardReset.info

How to Start Remote Control Computer and Screen Mirroring in SAMSUNG A310F Galaxy A3 (2016)? - HardReset.info

Mirror View Smart Flip Case For Samsung Galaxy A3 2017 A32017 SM A320F A320 Luxury original Magnetic fundas Leather Phone Cover|Flip Cases| - AliExpress

GENERIC Mirror Screen Protector compatible for Samsung Galaxy A3 Core with full coverage : Amazon.in: Electronics

How to Start Remote Control Computer and Screen Mirroring in SAMSUNG A310F Galaxy A3 (2016)? - HardReset.info