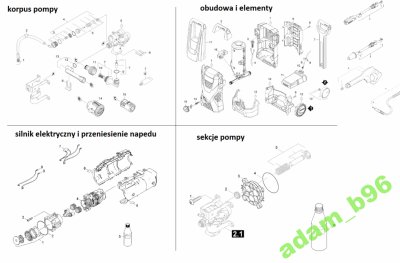
Głowica pompa do myjki Karcher K2 K3 zawór chemii zaworki uszczelniacze Full Control | sklep.techkar - Głowice, prowadnice tłoków - Części zamienne | sklep.techkar.pl
MYJKA CIŚNIENIOWA KARCHER K2 BASIC CAR PREMIUM (MYJKA CIŚNIENIOWA KARCHER K2 BASIC CAR x) • Cena, Opinie 8591678855 • Allegro.pl
Myjka ciśnieniowa Karcher K2 Basic 1.673-159.0 (1.673-159.0) • Cena, Opinie 12574811502 • Allegro.pl

Myjka ciśnieniowa KARCHER K 2 Basic 1.673-155.0 | Myjki ciśnieniowe | Hurtownia sklep z częściami zamiennymi do AGD Czesciagd.pl

Myjka ciśnieniowa KARCHER K2 Universal Edition 1.673-000.0 cena, opinie, dane techniczne | sklep internetowy Electro.pl