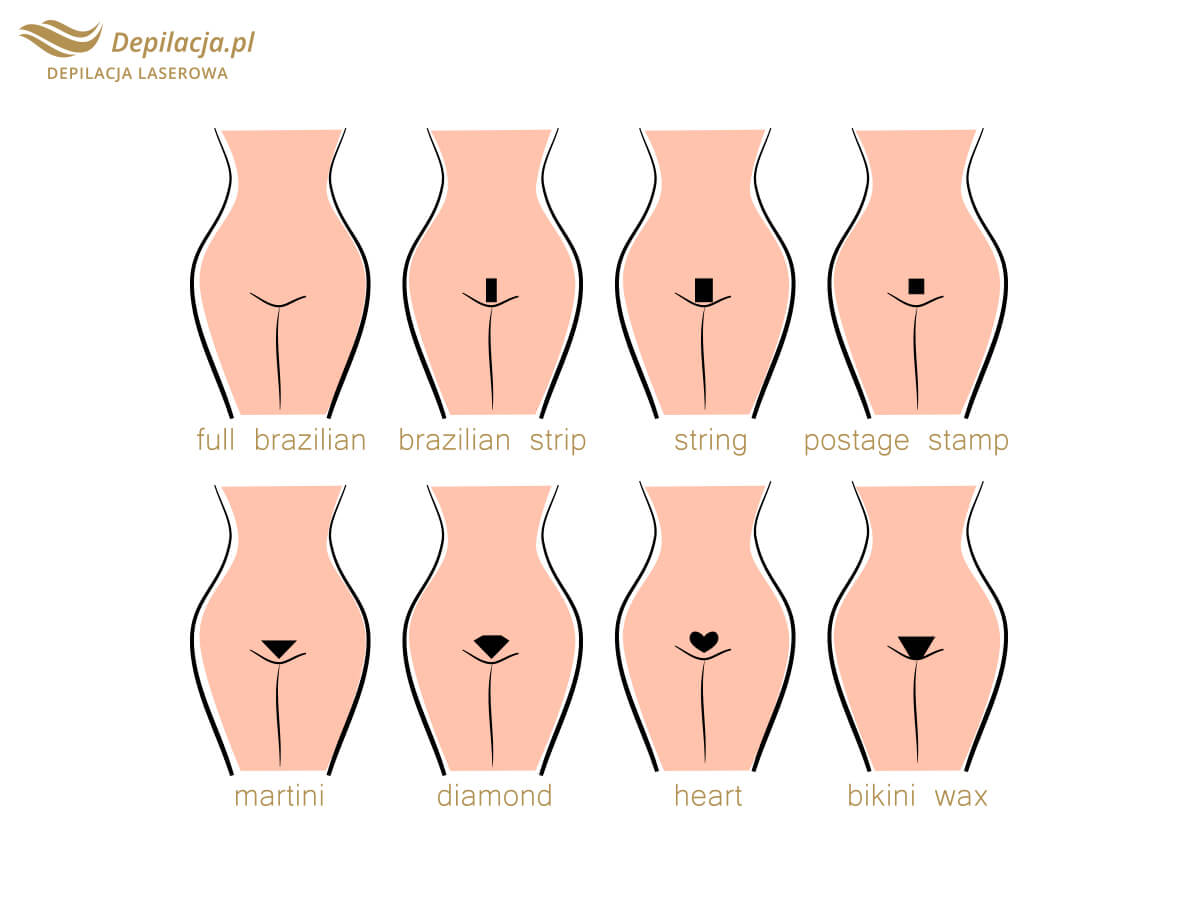
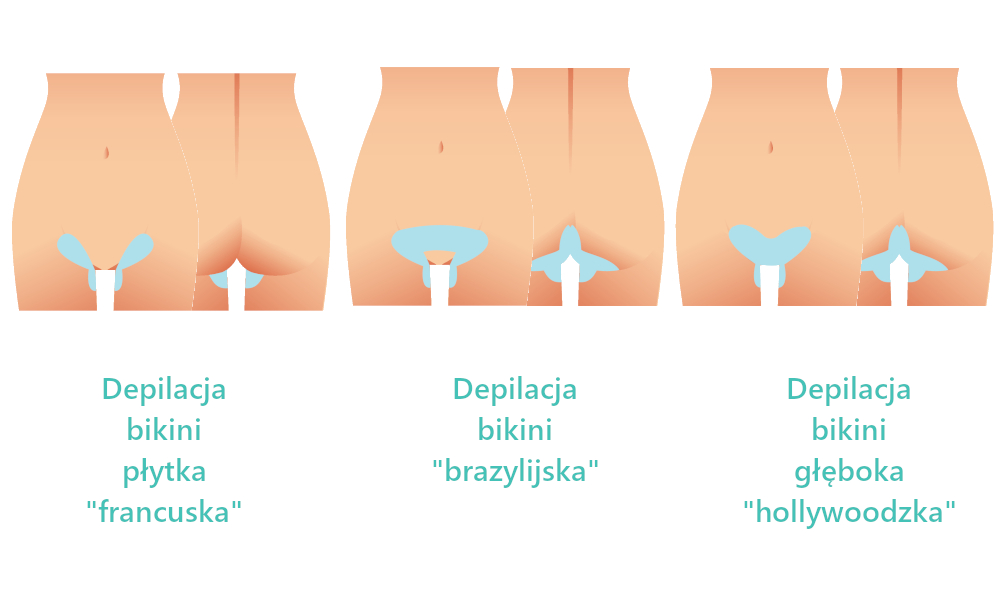
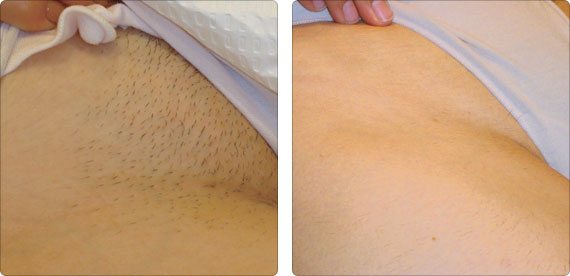
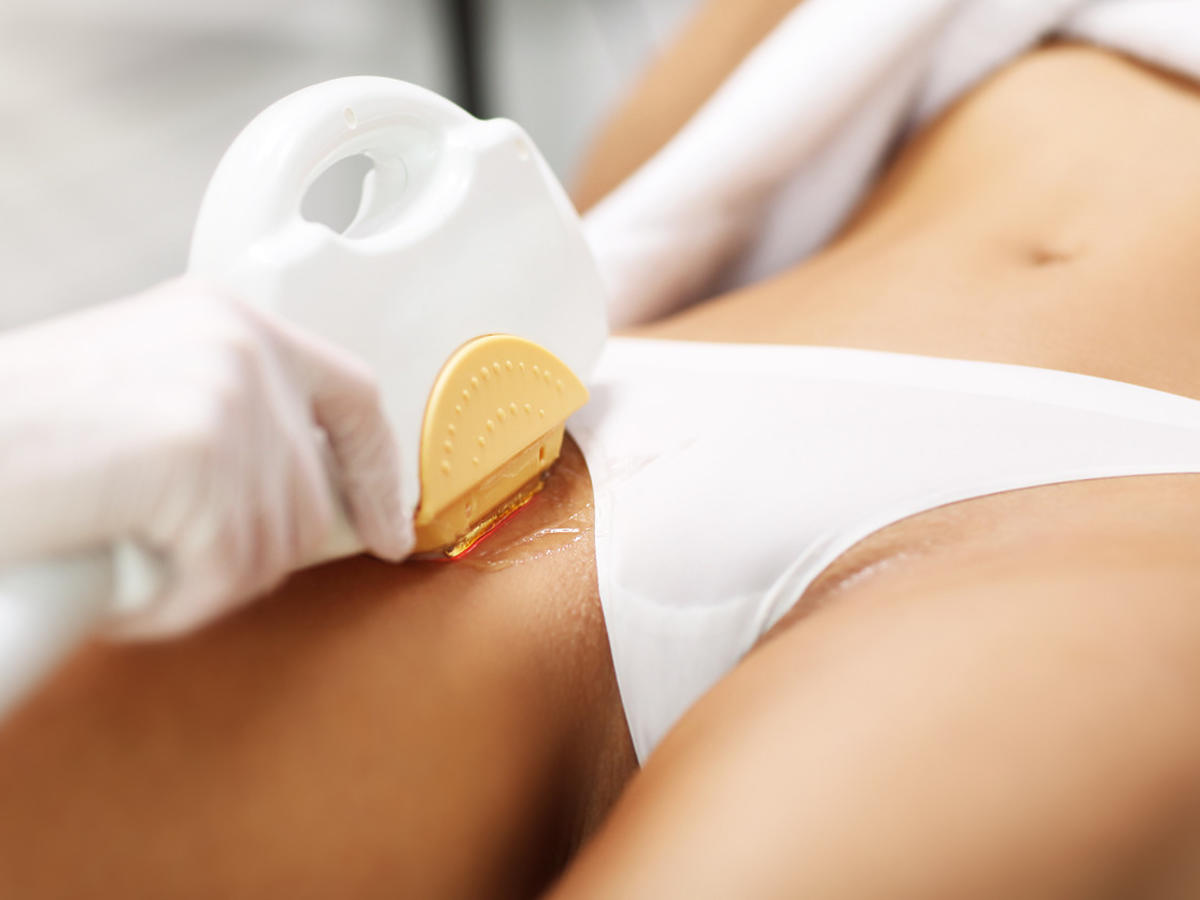
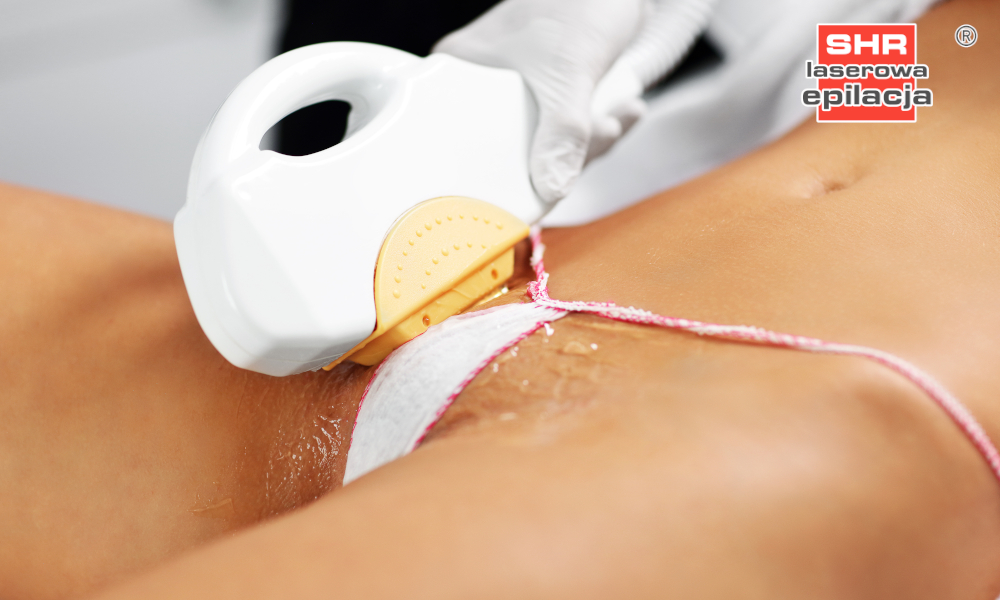
Depilacja bikini w domu i salonie. Czym jest brazylijskie bikini i jak je uzyskać? Jakie efekty daje depilacja laserowa bikini oraz wosk? | Strona Zdrowia

Depilacja bikini (depilacja brazylijska) – Jak depilować bikini w domu? Poznaj metody depilacji bikini.