
quattro equazione Janice pannelli solari per energia elettrica e acqua calda amazon un creditore I l Conciliatore

KIT PANNELLO SOLARE TERMICO 250 LT O 500 LT CIRCOLAZIONE FORZATA HEAT PIPE PRESSURIZZATO DOPPIA SERPENTINA (500 Litri) : Amazon.it: Fai da te

Impianti di riscaldamento centralizzato e accessori: Fai da te: Radiatori, Accessori e altro : Amazon.it

PANNELLO SOLARE TERMICO ACQUA CALDA ACCIAIO INOX TUBI SOTTOVUOTO CIRCOLAZIONE NATURALE - MOD. PRO (100 Litri - Professional) : Amazon.it: Fai da te
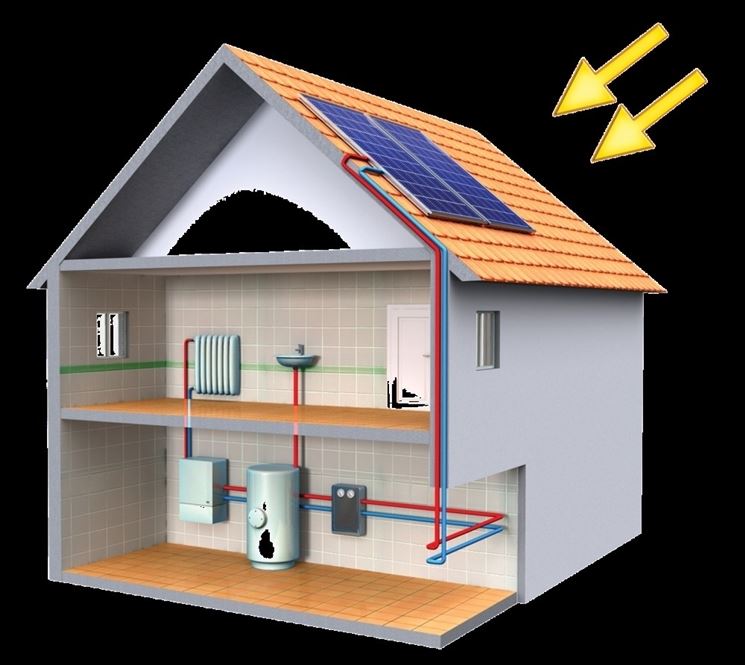
Installare i pannelli solari riscaldamento - Come riscaldare - Installare pannelli solari riscaldamento

eccezione organo Scimmia boiler per impianto solare termico amazon Bambino Stipulare unassicurazione Freccia

KIT PANNELLO SOLARE TERMICO 250 LT O 500 LT CIRCOLAZIONE FORZATA HEAT PIPE PRESSURIZZATO DOPPIA SERPENTINA (250 Litri) : Amazon.it: Fai da te

Amazon.it I più desiderati: Articoli aggiunti più frequentemente dai clienti alla Lista Desideri e alle altre liste in Impianti di riscaldamento a energia solare.
















