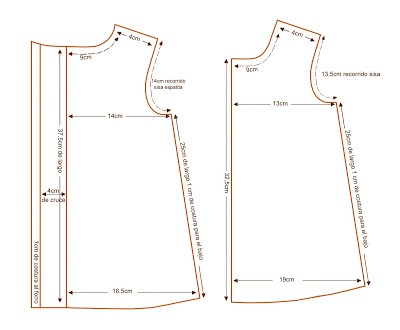
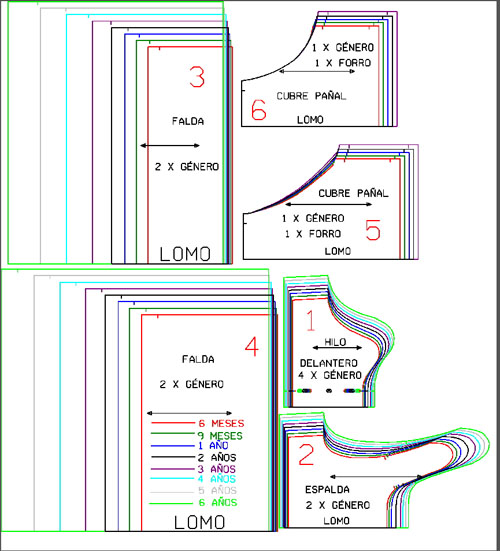
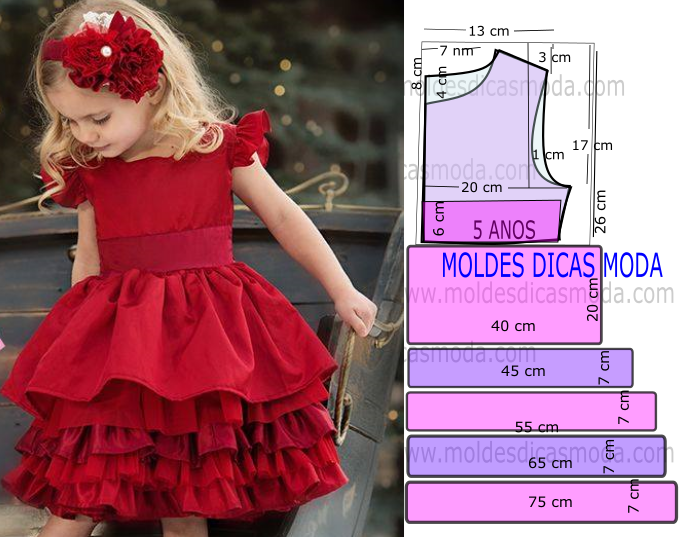
Diseños De Patrones Bordados A Mano,Vestido De Bebé Para Niña De O-4 Años Buy Mano Bordado Diseños Para Bebé Vestido De Niña De Patrones De Bordado A Mano Bebé Vestido Product

Fossen Niña Ropa Verano Vestido para 2-7 AñOs - Patrones De Personaje De Dibujos Animados - V Cuello Cordones En Espalda (3 años, Blanco): Amazon.es: Moda