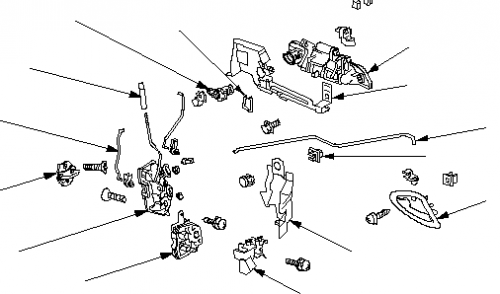
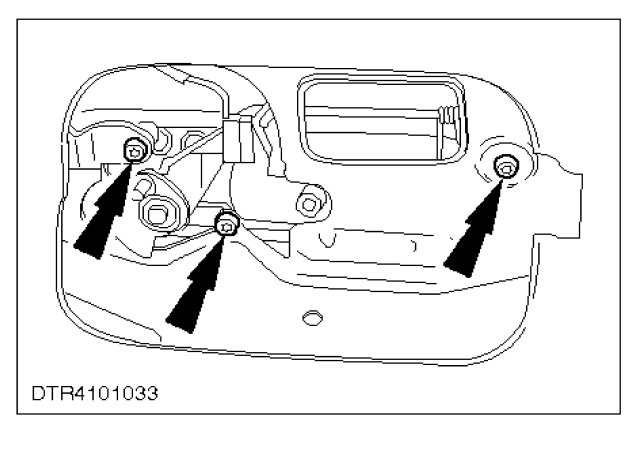
Honda Civic VI Liftback - Centralny zamek STIG, kluczyk scyzoryk - podłączenie. Honda Civic VI liftback • Blog auta • autoWcentrum.pl

Porady fachowca - Naprawa stacyjki HONDA, CIVIC, ACCORD , CRV 2002 -2013 - Dorabianie Kluczy i Programowanie Pilotów Samochodowych