
asrock h61-hvs quạt nguồn và quạt cpu quay vài giây rồi tắt rồi quay tiếp diễn liên tục màn hình đen - Asrock mainboard repair - Diễn đàn Kỹ Thuật Phần Cứng

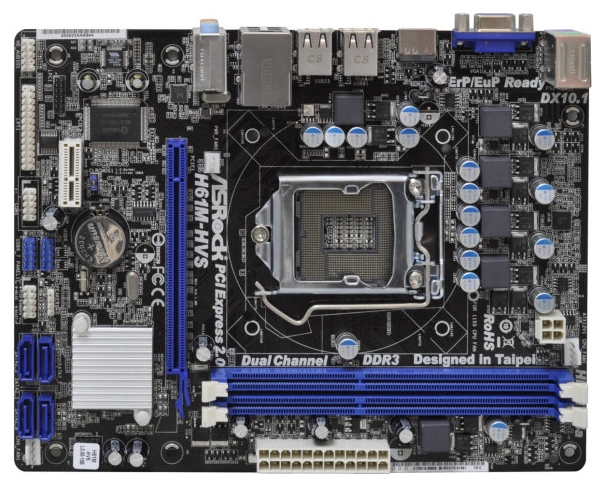
Amazon.com: ASRock H61M-HVS LGA1155/ Intel H61/ DDR3/ A&L/MicroATX Motherboard (Bulk) H61M-HVS Bulk White Box : Electronics

Amazon.com: ASRock H61M-HVS LGA1155/ Intel H61/ DDR3/ A&L/MicroATX Motherboard (Bulk) H61M-HVS Bulk White Box : Electronics

Asrock H61M-HVS Motherboard is not turn on. Problem with the BIOS. Motherboard black screen. - YouTube

Original Asrock H61M HVS Desktop Motherboard H61 Socket LGA 1155 For i3 i5 i7 DDR3 16G Micro ATX 100% Fully Test|desktop motherboard|motherboard h61lga 1155 - AliExpress

Amazon.com: ASRock H61M-HVS LGA1155/ Intel H61/ DDR3/ A&L/MicroATX Motherboard (Bulk) H61M-HVS Bulk White Box : Electronics












![ASRock H61M-DGS BIOS [PCAXE.COM] - YouTube ASRock H61M-DGS BIOS [PCAXE.COM] - YouTube](https://i.ytimg.com/vi/83Gsjp2FL6Q/maxresdefault.jpg)

.png)