
BEBC on Twitter: "Today's Phrase. 'Ball is in your court' Meaning - It is up to you to make the next decision or step https://t.co/gEH14J1HAg" / Twitter

The Ball Is In Your Court: Embracing Your Child's Dreams: Chalmers, Almarie: 9781599322131: Amazon.com: Books

Talk2Me English - This idiom ,'The ball is in your court' simply means that you have the opportunity to do what you think or feel. Idioms are a great way to express

Talaera - Idiom of the week: **The ball is in your court** Meaning: It's your turn to make a decision or do something #idioms #Englishvocabulary #learnEnglish https://hubs.ly/H0jmG-y0 | Facebook

Michael Scott Our Balls Are In Your Court GIF - Michael Scott Our Balls Are In Your Court Theoffice - Discover & Share GIFs







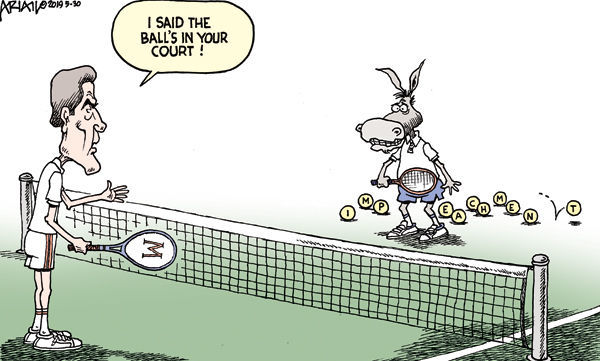



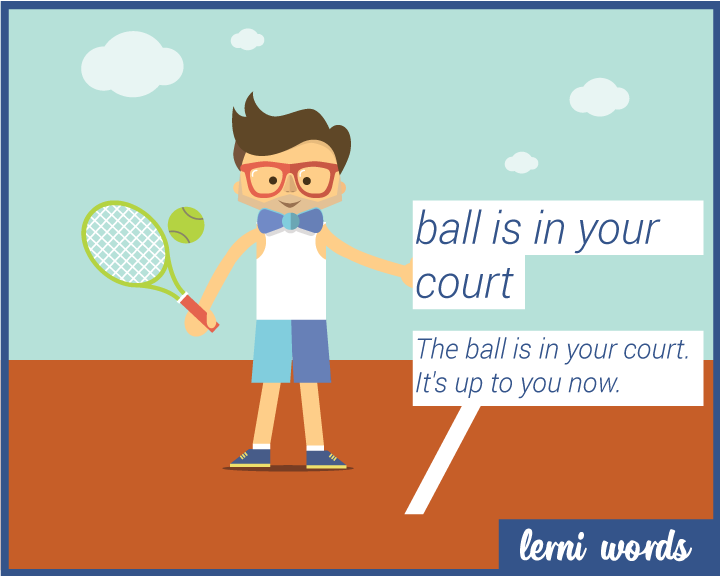







![OC] IDIOMS - THE BALL IS IN YOUR COURT : r/vocabulary OC] IDIOMS - THE BALL IS IN YOUR COURT : r/vocabulary](https://i.redd.it/82r2buia7ii71.png)