D61E.01/41 - Gniazdo komputerowe pojedyncze ekranowane RJ45 kategoria 6, z przesłoną przeciwkurzową (moduł) kremowy

SIMON BASIC Gniazdo komputerowe podwójne RJ45 kategoria 6 z przesłoną przeciwkurzową (moduł) czekoladowy mat metalizowany - sklep elektryczny el12

Kontakt-Simon Basic Moduł Gniazdo Komputerowe Rj45 Kategoria 6 Podwójne Z Przesłoną Przeciwkurzową . Montaż Na Łapki Lub Wkręty Inox B Bm62.01/21 – ABK Soft

SIMON BASIC Gniazdo komputerowe podwójne RJ45 kategoria 6, z przesłoną przeciwkurzową (moduł) srebrny mat, metalizowany - sklep elektryczny el12

Voice and data flat plate with dust cover for 1 element and 1 RJ45 connector category 6 UTP white Simon K45 | SIMON
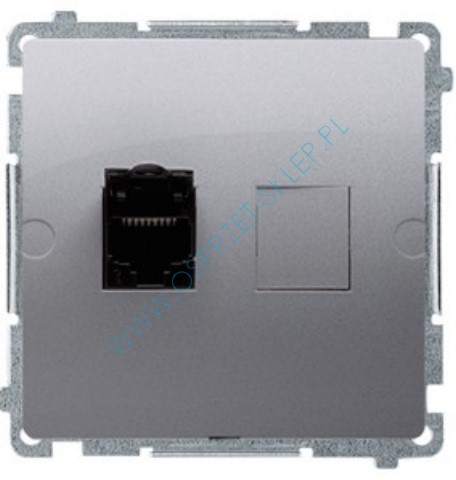
Simon Basic Gniazdo komputerowe pojedyncze RJ45 kat.5e satynowe BMF51.02/29 – Kontakt-Simon | TIM SA

Kontakt-Simon Basic Moduł Gniazdo Komputerowe Rj45 Kat.5E + Telefoniczne Rj12 . Montaż Gniazda Na Wkręty Do Puszki Białe Bm5T.02/11 – ABAcosunwejherowo

Flat voice and data plate without dust cover for 1 RJ45 3M® for 1 element white Simon 500 Cima | SIMON
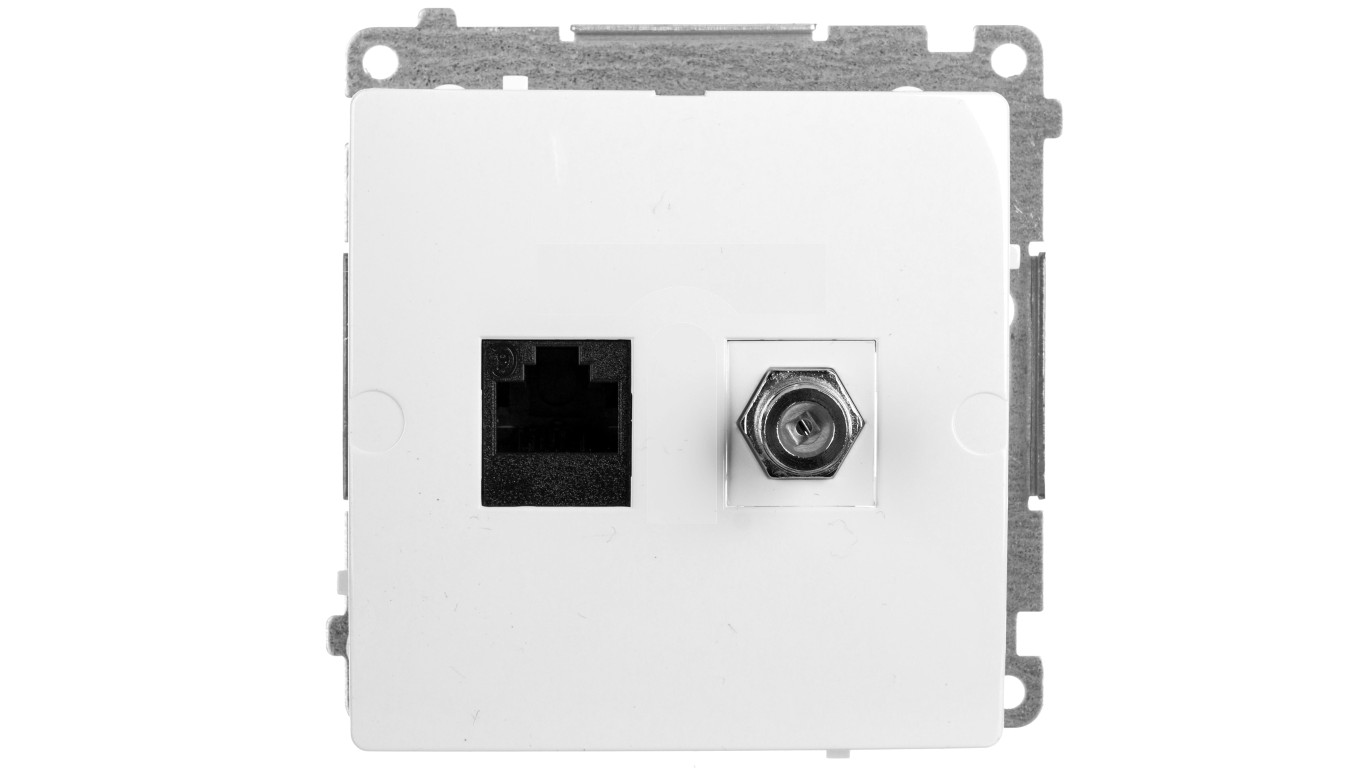
Simon Basic Gniazdo antenowe typu F + komputerowe RJ45 kat.6 białe BMAFRJ45.01/11 – Kontakt-Simon | TIM SA

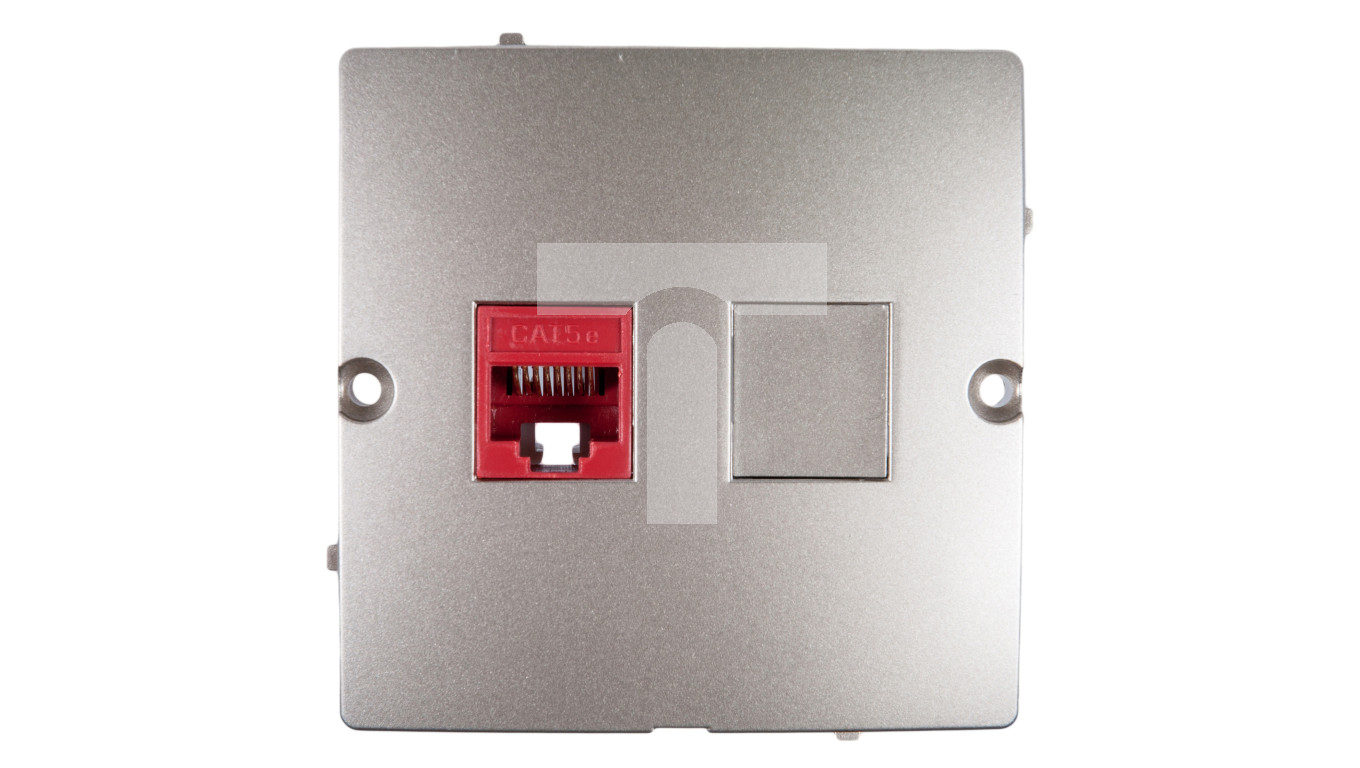
Simon Basic Gniazdo komputerowe podwójne RJ45 kat. 5e srebrny mat BMF52.02 43 - Gniazda teleinformatyczne - Gniazda - Gniazda i łączniki