150/6 cal zmienna częstotliwość wentylator kanałowy kuchnia urządzenie do usuwania oparów wc potężny cichy wentylator wymiana powietrza wentylator|Wentylatory wyciągowe| - AliExpress

Soler&Palau Ultra cichy wentylator kanałowy TD 500/150-160 CILENT S&P - 6" - do 550 m3/h : Amazon.pl: Narzędzia i renowacja domu

PROFAN - fi150mm, 2-BIEGI 405/520m3/h - CICHY WENTYLATOR KANAŁOWY Z KABLEM w atrakcyjnej cenie! | Growshop domowauprawa.pl

HG Power 150 mm, regulowany wentylator wylotowy, wentylator rurowy z wentylatorem, regulatorem prędkości obrotowej, energooszczędny, cichy wentylator kanałowy, wentylator łazienkowy, wentylator rurowy do kuchni, łazienki, garażu, dmuchawa wyciągowa ...

Wentylator kanałowy TNI SILENT 150-160 dwubiegowy do wentylacji pomieszczeń o niskim stopniu zapylenia z przepustnicą zwrotną cichy z wysokiej jakości tworzywa sztucznego 530/410 m3/h Klivago

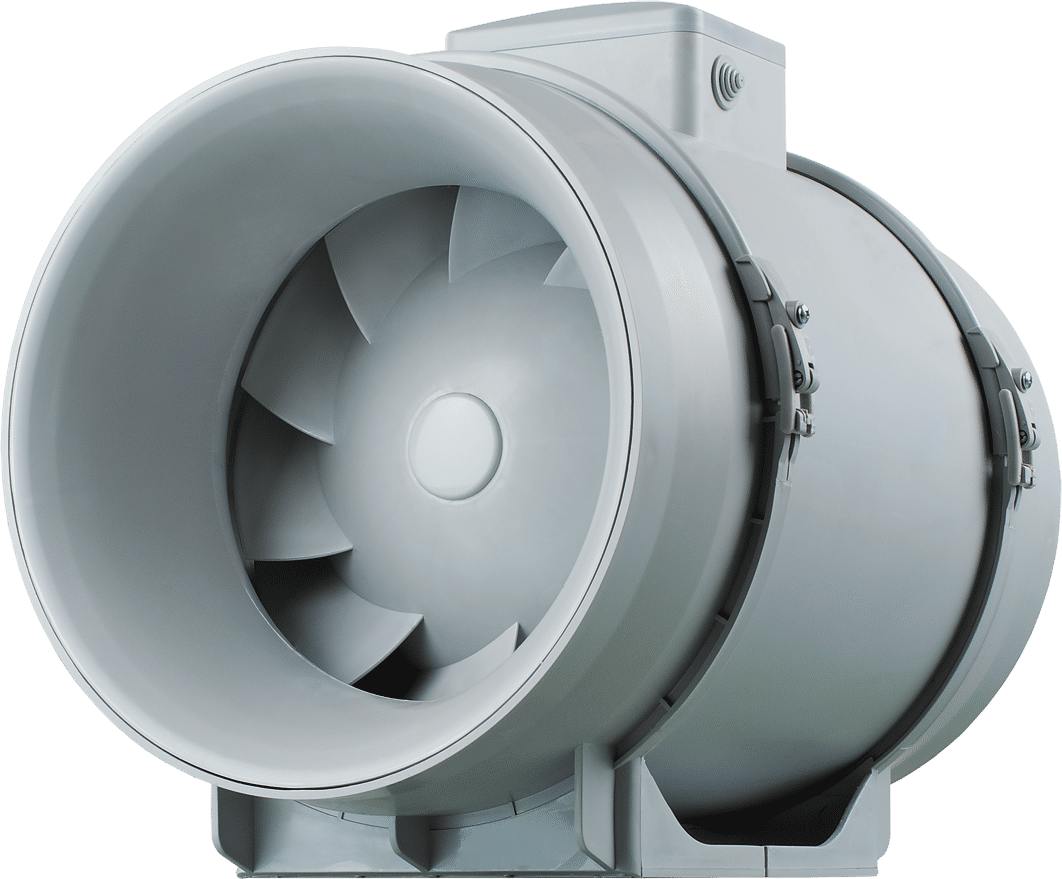
Wentylator kanałowy TT SILENT-M 160 cichy niskoszumowy izolowany dwubiegowy do kanałów wentylacyjnych okrągłych metalowy malowany proszkowo 405/555 m3/h Klivago

Wentylator łazienkowy Bestfan Cichy Wentylator Kanałowy fi 150mm BHF-150 BHF150 - Opinie i ceny na Ceneo.pl

PROFAN - fi100mm, 2-BIEGI 145/187m3/h - CICHY WENTYLATOR KANAŁOWY Z KABLEM w atrakcyjnej cenie! | Growshop domowauprawa.pl