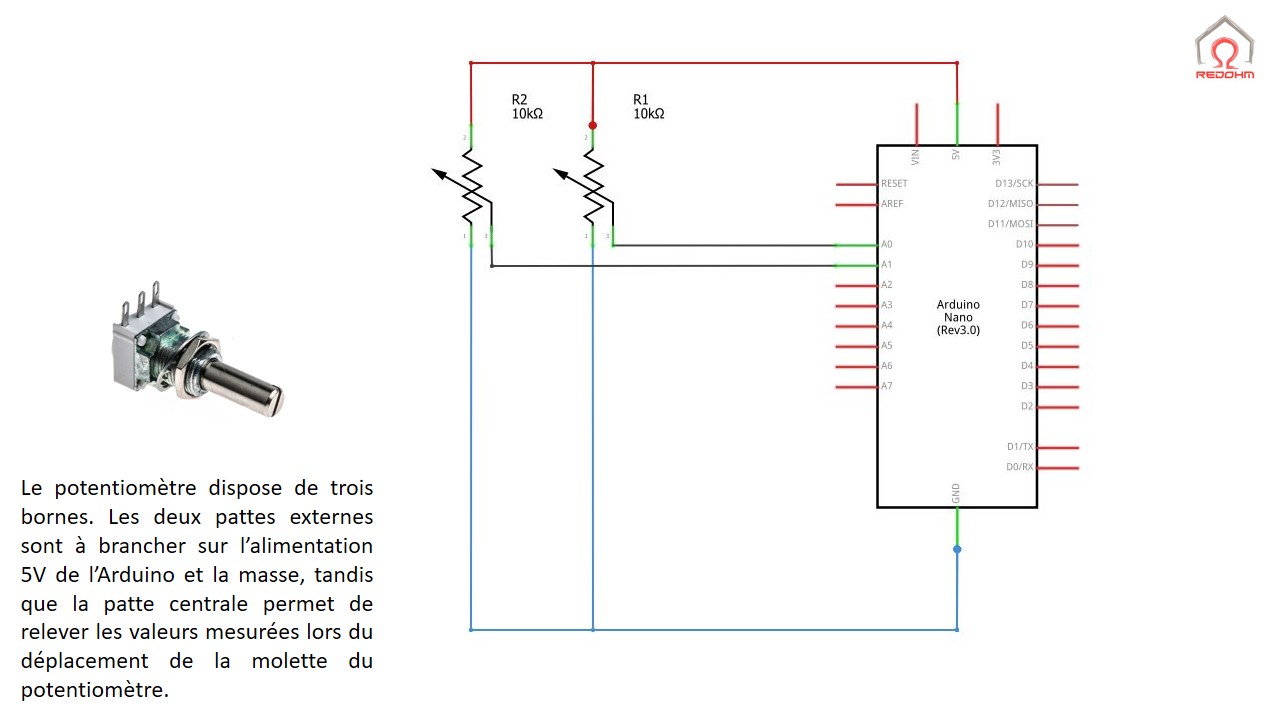
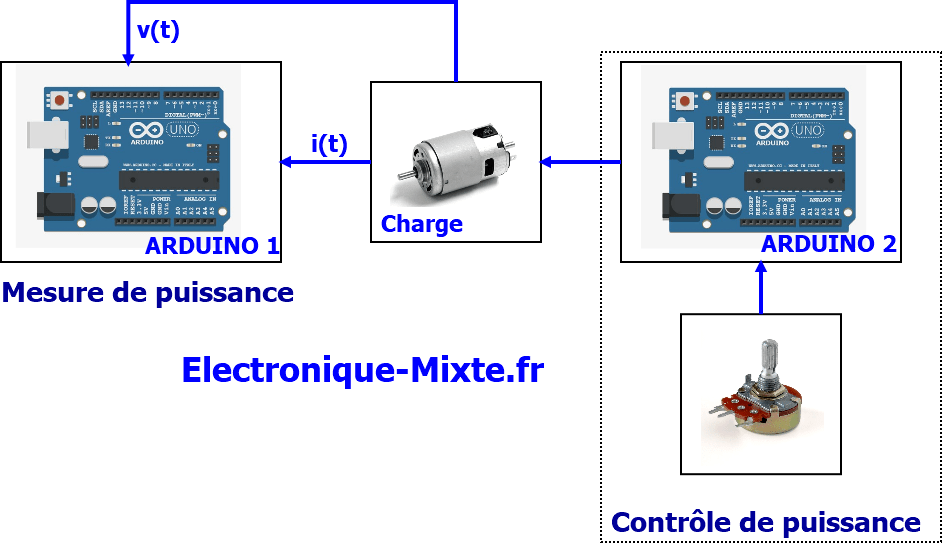
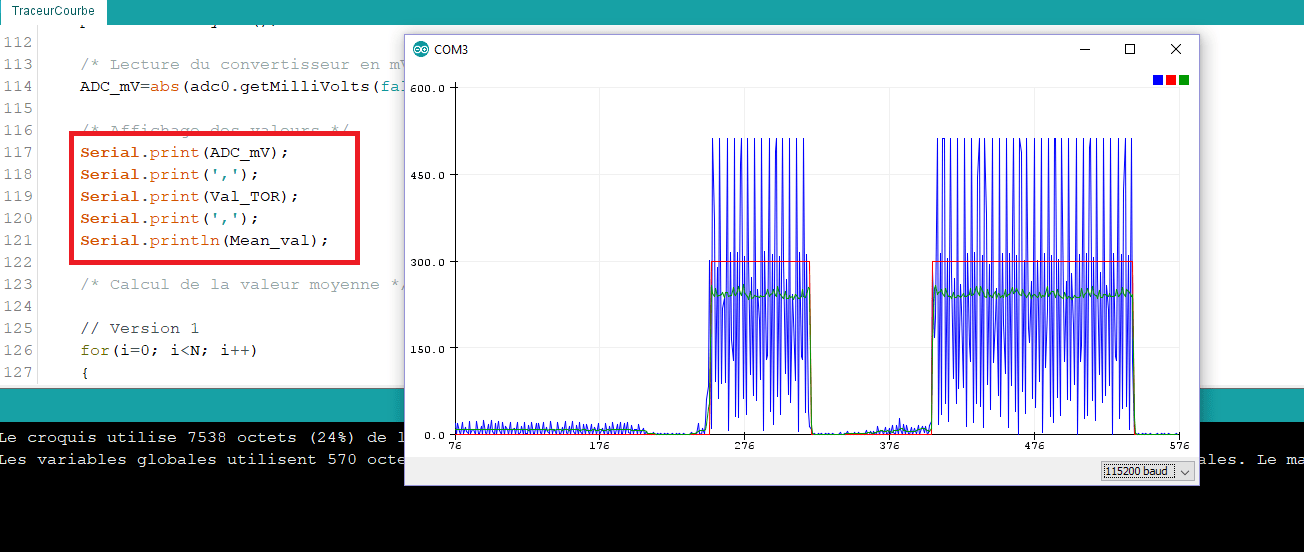
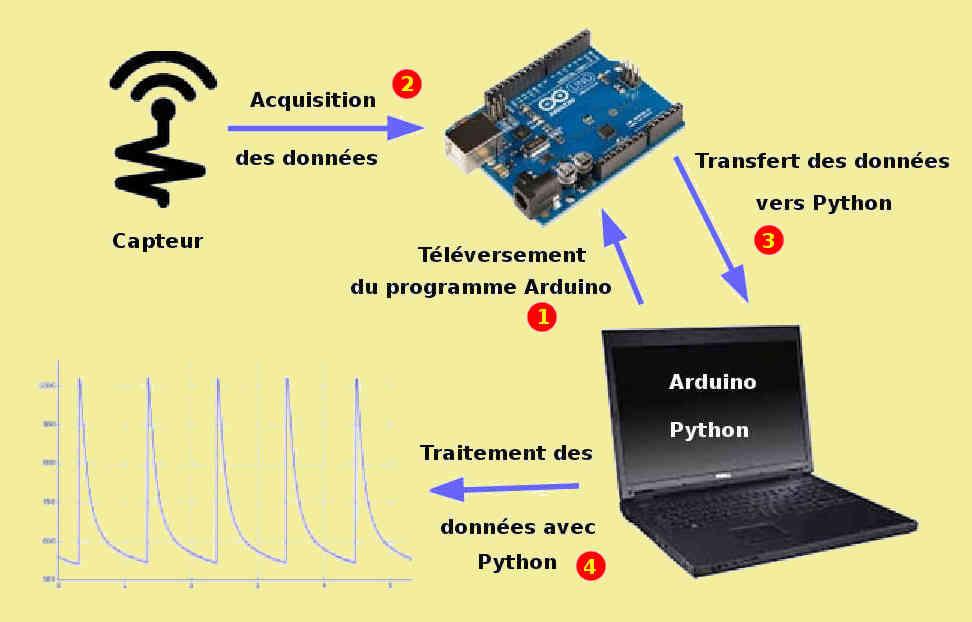
MQ-2. Débuter avec le détecteur de gaz et fumées avec du code Arduino. Calibration, concentration en ppm • Domotique et objets connectés à faire soi-même
Tutoriels pour Arduino • Afficher le sujet - Capteur d'humidité du sol (analogique et numérique) avec Ard

La conversion analogique / numérique avec Arduino / Genuino | Carnet du maker - L'esprit Do It Yourself

Module de capteur photosensible à résistance optique, 3 broches, détection de lumière pour arduino, Kit de bricolage KY 018 KY018 | AliExpress

Module de capteur photosensible à résistance optique, 3 broches, détection de lumière pour arduino, Kit de bricolage KY 018 KY018 | AliExpress